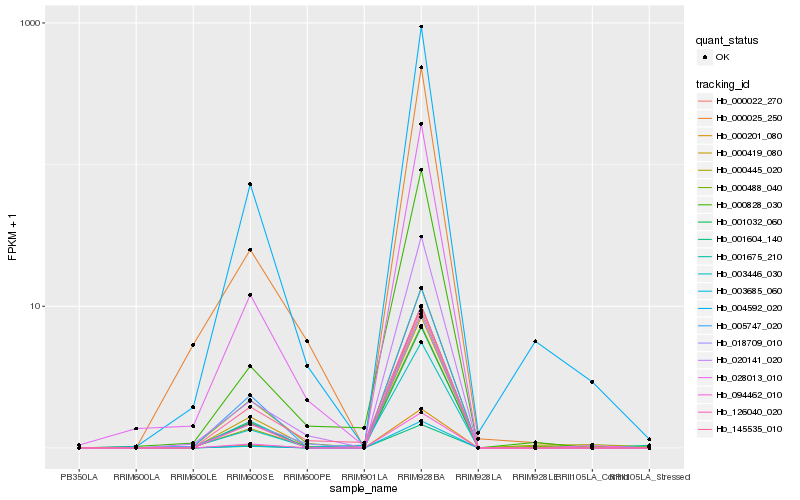
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003446_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 2 |
Hb_001032_060 |
0.0818819408 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 3 |
Hb_028013_010 |
0.082448084 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_145535_010 |
0.0876318807 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 5 |
Hb_018709_010 |
0.0897000575 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 6 |
Hb_000419_080 |
0.0940909805 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 7 |
Hb_000445_020 |
0.0961831701 |
- |
- |
GABA-specific permease, putative [Ricinus communis] |
| 8 |
Hb_001675_210 |
0.0973950894 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 9 |
Hb_000488_040 |
0.0979401354 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000828_030 |
0.0995048792 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 11 |
Hb_000025_250 |
0.0997709252 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 12 |
Hb_004592_020 |
0.1003062174 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 13 |
Hb_094462_010 |
0.1007539943 |
- |
- |
hypothetical protein CISIN_1g033698mg [Citrus sinensis] |
| 14 |
Hb_005747_020 |
0.1010440579 |
- |
- |
bacterial-induced peroxidase [Gossypium schwendimanii] |
| 15 |
Hb_000022_270 |
0.1016389342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003685_060 |
0.1021517749 |
- |
- |
PREDICTED: laccase-15 [Jatropha curcas] |
| 17 |
Hb_001604_140 |
0.1027917977 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |
| 18 |
Hb_000201_080 |
0.1040115429 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 19 |
Hb_126040_020 |
0.1055206577 |
- |
- |
- |
| 20 |
Hb_020141_020 |
0.1059377671 |
- |
- |
- |