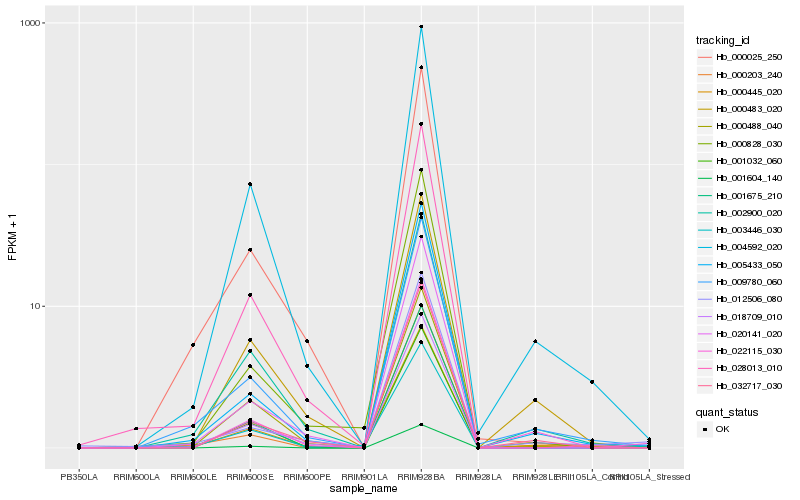
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_020 |
0.0 |
- |
- |
GABA-specific permease, putative [Ricinus communis] |
| 2 |
Hb_009780_060 |
0.0787673325 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_004592_020 |
0.0791271667 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 4 |
Hb_002900_020 |
0.0886383207 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 5 |
Hb_005433_050 |
0.0951293883 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 6 |
Hb_000488_040 |
0.0956959464 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003446_030 |
0.0961831701 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 8 |
Hb_000203_240 |
0.0977257494 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_012506_080 |
0.0983163157 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_000483_020 |
0.1018570039 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000828_030 |
0.104033735 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 12 |
Hb_022115_030 |
0.1066010173 |
- |
- |
PREDICTED: umecyanin-like [Jatropha curcas] |
| 13 |
Hb_032717_030 |
0.1074191113 |
- |
- |
PREDICTED: CASP-like protein 1B2 [Jatropha curcas] |
| 14 |
Hb_028013_010 |
0.1080687629 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_018709_010 |
0.110514225 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 16 |
Hb_001675_210 |
0.1116799067 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 17 |
Hb_001032_060 |
0.11174299 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 18 |
Hb_020141_020 |
0.1154747221 |
- |
- |
- |
| 19 |
Hb_000025_250 |
0.1159534016 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 20 |
Hb_001604_140 |
0.1182319397 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |