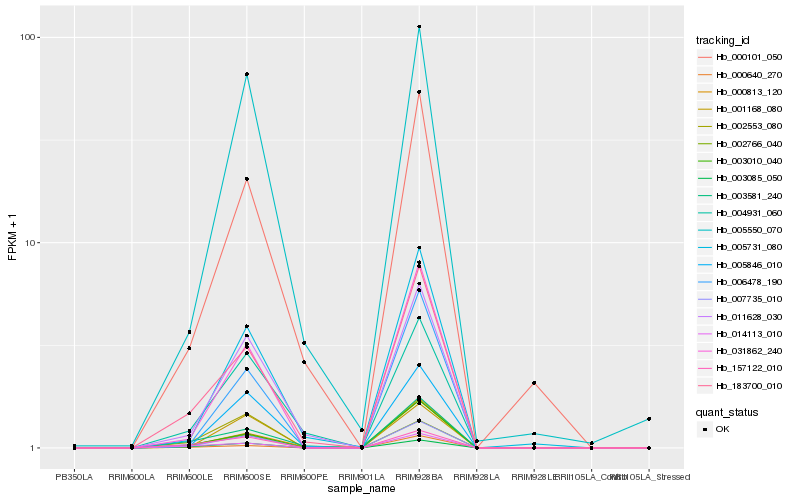
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002766_040 |
0.0 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 2 |
Hb_007735_010 |
0.0277305406 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 3 |
Hb_002553_080 |
0.0790414795 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF096-like [Eucalyptus grandis] |
| 4 |
Hb_183700_010 |
0.0886127364 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_003085_050 |
0.0906420242 |
- |
- |
Polygalacturonase non-catalytic subunit AroGP3 precursor, putative [Ricinus communis] |
| 6 |
Hb_014113_010 |
0.099105707 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_157122_010 |
0.0992393996 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 8 |
Hb_031862_240 |
0.114616164 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 9 |
Hb_011628_030 |
0.1209245946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003581_240 |
0.1305815556 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_005550_070 |
0.1344348422 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 12 |
Hb_005846_010 |
0.1345910098 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 13 |
Hb_004931_060 |
0.1391944495 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4 [Nelumbo nucifera] |
| 14 |
Hb_006478_190 |
0.1435842942 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 15 |
Hb_001168_080 |
0.1435935712 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_000813_120 |
0.1441780464 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Tarenaya hassleriana] |
| 17 |
Hb_000101_050 |
0.144792926 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_005731_080 |
0.1487560722 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 19 |
Hb_003010_040 |
0.1488541378 |
- |
- |
hypothetical protein AALP_AA7G248200 [Arabis alpina] |
| 20 |
Hb_000640_270 |
0.1508699357 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable pectinesterase 30 [Jatropha curcas] |