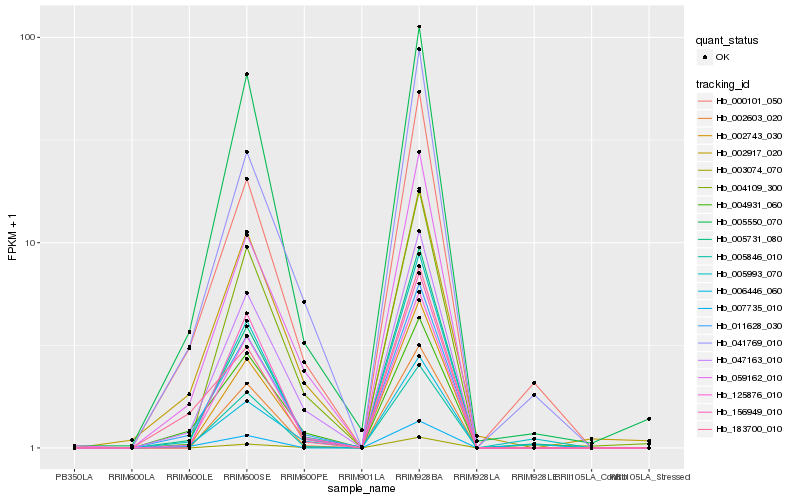
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011628_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_047163_010 |
0.0438575124 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 3 |
Hb_059162_010 |
0.0646647964 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_125876_010 |
0.0667203576 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_004931_060 |
0.0710424699 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4 [Nelumbo nucifera] |
| 6 |
Hb_005550_070 |
0.0715259366 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 7 |
Hb_002743_030 |
0.0770556584 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 8 |
Hb_005846_010 |
0.0787539669 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 9 |
Hb_005731_080 |
0.0946481448 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 10 |
Hb_004109_300 |
0.0959062845 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 11 |
Hb_005993_070 |
0.0977239606 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 12 |
Hb_000101_050 |
0.100189839 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_006446_060 |
0.1017451406 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 14 |
Hb_041769_010 |
0.106056586 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 15 |
Hb_002603_020 |
0.1075604973 |
- |
- |
PREDICTED: O-acetyl-ADP-ribose deacetylase MACROD2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002917_020 |
0.1077386516 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 17 |
Hb_156949_010 |
0.1099047182 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 18 |
Hb_007735_010 |
0.1101821649 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 19 |
Hb_183700_010 |
0.1158415852 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_003074_070 |
0.1178370911 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |