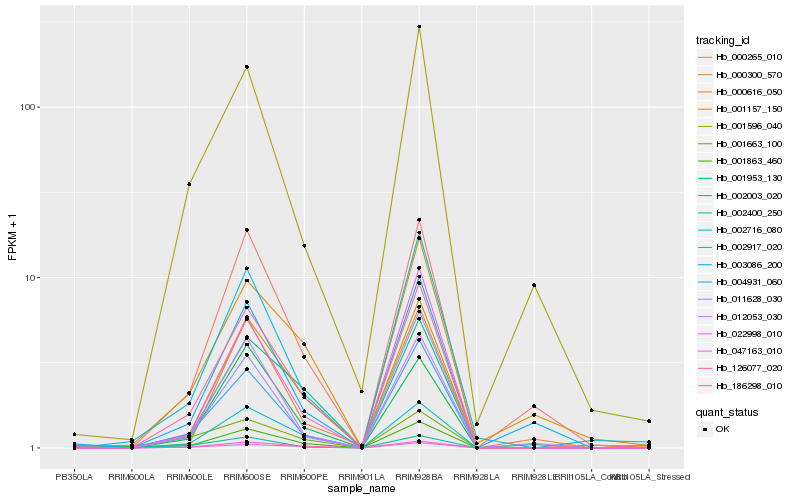
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186298_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_004931_060 |
0.0857391078 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4 [Nelumbo nucifera] |
| 3 |
Hb_126077_020 |
0.0985128884 |
- |
- |
Kinase family protein with leucine-rich repeat domain [Theobroma cacao] |
| 4 |
Hb_002917_020 |
0.1036726407 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 5 |
Hb_000265_010 |
0.1165369647 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_002716_080 |
0.1186003854 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 7 |
Hb_001863_460 |
0.1211197371 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 8 |
Hb_022998_010 |
0.1234848236 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_001157_150 |
0.1251015762 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 10 |
Hb_002003_020 |
0.1267274822 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 11 |
Hb_002400_250 |
0.1281029544 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_000300_570 |
0.1283435538 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 13 |
Hb_012053_030 |
0.1325145246 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g22810 [Jatropha curcas] |
| 14 |
Hb_003086_200 |
0.1336331891 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 15 |
Hb_047163_010 |
0.1337818957 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 16 |
Hb_001596_040 |
0.1346192248 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase 1-like [Populus euphratica] |
| 17 |
Hb_001663_100 |
0.1366329058 |
- |
- |
- |
| 18 |
Hb_000616_050 |
0.1387727337 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 19 |
Hb_011628_030 |
0.1389668581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001953_130 |
0.1395651444 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |