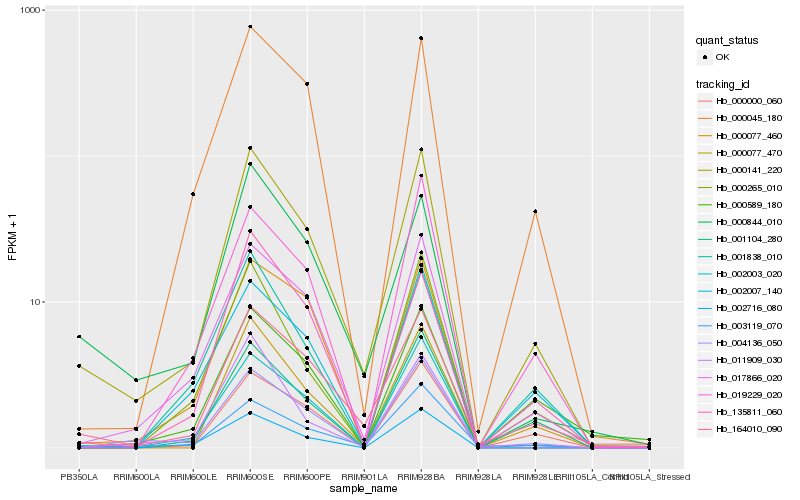
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000141_220 |
0.0 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 2 |
Hb_164010_090 |
0.0976780528 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 3 |
Hb_017866_020 |
0.1095926773 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 4 |
Hb_002003_020 |
0.1156516309 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 5 |
Hb_000077_460 |
0.1331566722 |
- |
- |
- |
| 6 |
Hb_000844_010 |
0.1362956699 |
- |
- |
PREDICTED: lysine histidine transporter-like 8 [Jatropha curcas] |
| 7 |
Hb_000045_180 |
0.1367989966 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 8 |
Hb_000000_060 |
0.1373912062 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 9 |
Hb_003119_070 |
0.1377063291 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_001104_280 |
0.1409860513 |
- |
- |
PREDICTED: UPF0481 protein At3g47200 [Jatropha curcas] |
| 11 |
Hb_002007_140 |
0.1412129369 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 12 |
Hb_002716_080 |
0.1429707703 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 13 |
Hb_001838_010 |
0.1453111146 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |
| 14 |
Hb_011909_030 |
0.1494200977 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |
| 15 |
Hb_135811_060 |
0.1519680208 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 16 |
Hb_000265_010 |
0.1532746719 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_000077_470 |
0.1532763597 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 18 |
Hb_004136_050 |
0.1539643048 |
- |
- |
- |
| 19 |
Hb_000589_180 |
0.1540592461 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 20 |
Hb_019229_020 |
0.1541264722 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |