| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
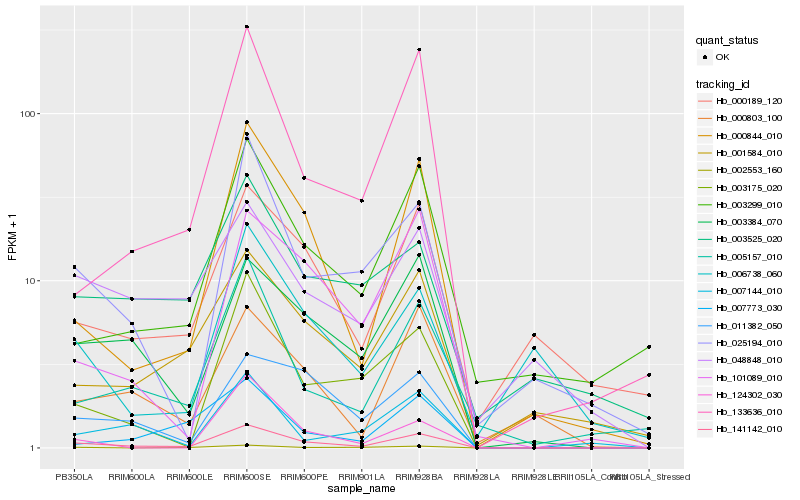
Hb_141142_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_21320 [Jatropha curcas] |
| 2 |
Hb_000844_010 |
0.1523169894 |
- |
- |
PREDICTED: lysine histidine transporter-like 8 [Jatropha curcas] |
| 3 |
Hb_003384_070 |
0.1726285903 |
- |
- |
PREDICTED: methylesterase 17-like [Populus euphratica] |
| 4 |
Hb_003175_020 |
0.1847961859 |
- |
- |
homeobox protein knotted-1, putative [Ricinus communis] |
| 5 |
Hb_025194_010 |
0.1927622988 |
- |
- |
PREDICTED: uncharacterized protein LOC105644857 [Jatropha curcas] |
| 6 |
Hb_133636_010 |
0.1950889741 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 7 |
Hb_005157_010 |
0.1960169717 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 8 |
Hb_011382_050 |
0.196672461 |
- |
- |
hypothetical protein PRUPE_ppa025935mg [Prunus persica] |
| 9 |
Hb_101089_010 |
0.199591375 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 10 |
Hb_002553_160 |
0.2008679528 |
- |
- |
PREDICTED: uncharacterized protein LOC104590935 [Nelumbo nucifera] |
| 11 |
Hb_001584_010 |
0.2015879964 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 12 |
Hb_000803_100 |
0.2033769477 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 13 |
Hb_003299_010 |
0.207243539 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 14 |
Hb_000189_120 |
0.2126790392 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 15 |
Hb_007773_030 |
0.2240443819 |
- |
- |
- |
| 16 |
Hb_124302_030 |
0.2278532356 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_048848_010 |
0.2282920864 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 18 |
Hb_006738_060 |
0.2288674929 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_007144_010 |
0.2295108642 |
- |
- |
- |
| 20 |
Hb_003525_020 |
0.2309356042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |