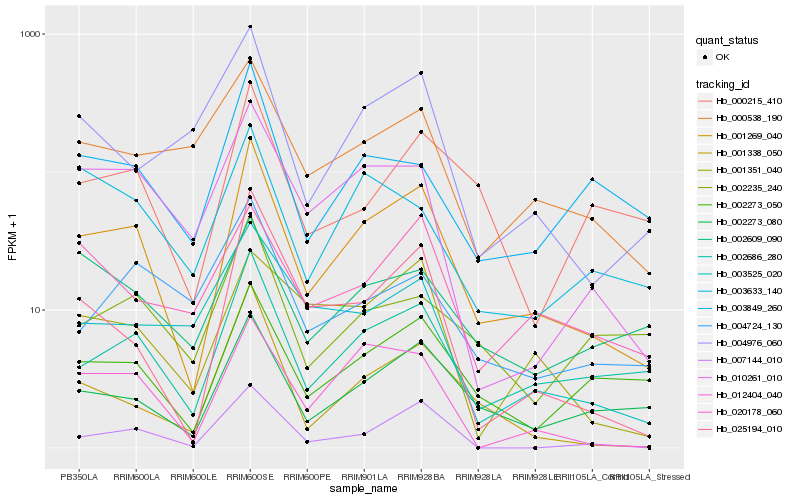
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_040 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_002686_280 |
0.1463694621 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_002273_080 |
0.1646693678 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 4 |
Hb_002273_050 |
0.1775018021 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_012404_040 |
0.1785531898 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 6 |
Hb_001338_050 |
0.1788123297 |
- |
- |
hypothetical protein POPTR_0016s05400g [Populus trichocarpa] |
| 7 |
Hb_025194_010 |
0.1796695536 |
- |
- |
PREDICTED: uncharacterized protein LOC105644857 [Jatropha curcas] |
| 8 |
Hb_010261_010 |
0.1893475577 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 9 |
Hb_007144_010 |
0.191646923 |
- |
- |
- |
| 10 |
Hb_000215_410 |
0.2065134716 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 11 |
Hb_004976_060 |
0.2066016938 |
- |
- |
- |
| 12 |
Hb_000538_190 |
0.2071813312 |
- |
- |
PREDICTED: protein DJ-1 homolog D [Jatropha curcas] |
| 13 |
Hb_003849_260 |
0.2094470779 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 14 |
Hb_003525_020 |
0.2131453903 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001351_040 |
0.214044396 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_002609_090 |
0.2173820446 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_004724_130 |
0.2181688504 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 18 |
Hb_002235_240 |
0.2182968347 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 19 |
Hb_003633_140 |
0.2189464658 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 20 |
Hb_020178_060 |
0.2193679548 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |