| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
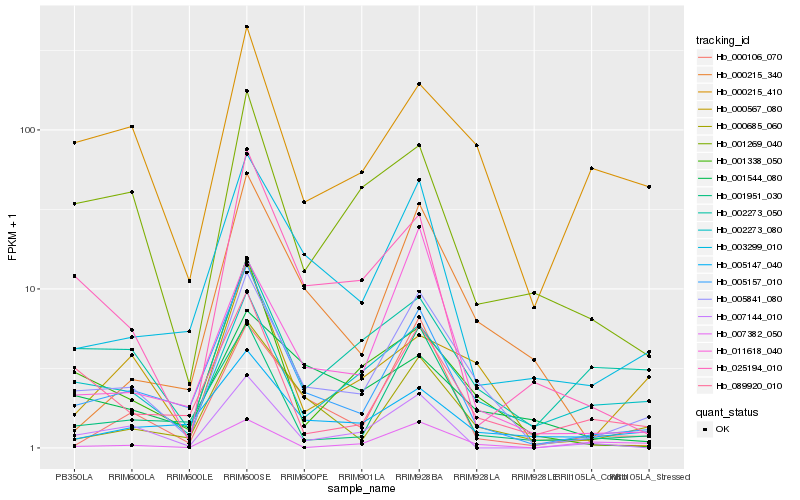
Hb_005841_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s11370g [Populus trichocarpa] |
| 2 |
Hb_005157_010 |
0.1608016189 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 3 |
Hb_000685_060 |
0.1886515091 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_003299_010 |
0.1895172596 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 5 |
Hb_002273_080 |
0.1912831332 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 6 |
Hb_000215_410 |
0.2040519503 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 7 |
Hb_000106_070 |
0.207535435 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 8 |
Hb_001951_030 |
0.2075787841 |
- |
- |
hypothetical protein JCGZ_04584 [Jatropha curcas] |
| 9 |
Hb_011618_040 |
0.2084878572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_089920_010 |
0.2127635874 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_000567_080 |
0.2182620522 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 12 |
Hb_001338_050 |
0.2187840493 |
- |
- |
hypothetical protein POPTR_0016s05400g [Populus trichocarpa] |
| 13 |
Hb_001269_040 |
0.2239160361 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_000215_340 |
0.2242039665 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 15 |
Hb_002273_050 |
0.2252088375 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_005147_040 |
0.2258211351 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 17 |
Hb_007144_010 |
0.2277355183 |
- |
- |
- |
| 18 |
Hb_007382_050 |
0.2343516445 |
- |
- |
PREDICTED: uncharacterized protein LOC105645009 [Jatropha curcas] |
| 19 |
Hb_025194_010 |
0.2355966022 |
- |
- |
PREDICTED: uncharacterized protein LOC105644857 [Jatropha curcas] |
| 20 |
Hb_001544_080 |
0.2360878186 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |