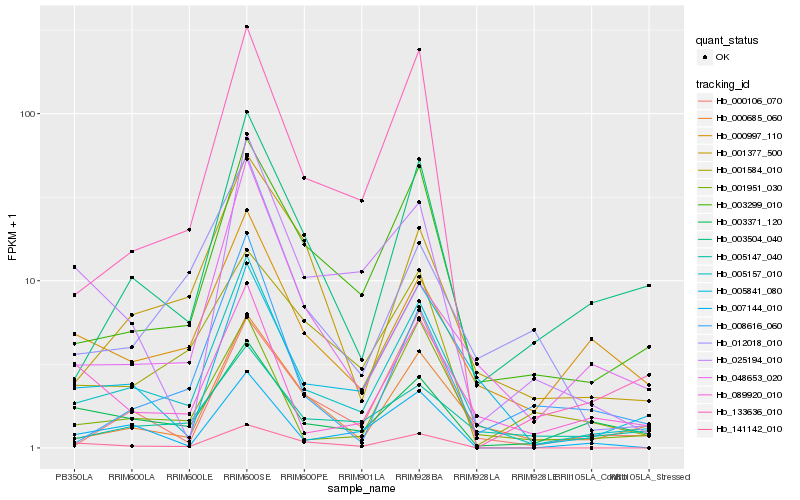
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005157_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 2 |
Hb_003299_010 |
0.1399769996 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 3 |
Hb_005841_080 |
0.1608016189 |
- |
- |
hypothetical protein POPTR_0018s11370g [Populus trichocarpa] |
| 4 |
Hb_000685_060 |
0.1628576799 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_001951_030 |
0.1695143605 |
- |
- |
hypothetical protein JCGZ_04584 [Jatropha curcas] |
| 6 |
Hb_003504_040 |
0.1705345434 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 7 |
Hb_133636_010 |
0.1706824982 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 8 |
Hb_005147_040 |
0.1899285385 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 9 |
Hb_001377_500 |
0.1919314083 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 10 |
Hb_003371_120 |
0.1920307115 |
- |
- |
hypothetical protein JCGZ_05278 [Jatropha curcas] |
| 11 |
Hb_089920_010 |
0.1930685227 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_007144_010 |
0.1958437817 |
- |
- |
- |
| 13 |
Hb_000997_110 |
0.1959591454 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 14 |
Hb_141142_010 |
0.1960169717 |
- |
- |
hypothetical protein JCGZ_21320 [Jatropha curcas] |
| 15 |
Hb_048653_020 |
0.2009745256 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_008616_060 |
0.20834708 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 17 |
Hb_001584_010 |
0.2090750423 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 18 |
Hb_025194_010 |
0.209453656 |
- |
- |
PREDICTED: uncharacterized protein LOC105644857 [Jatropha curcas] |
| 19 |
Hb_000106_070 |
0.2113079635 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 20 |
Hb_012018_010 |
0.2138422226 |
- |
- |
hypothetical protein MIMGU_mgv1a0024251mg, partial [Erythranthe guttata] |