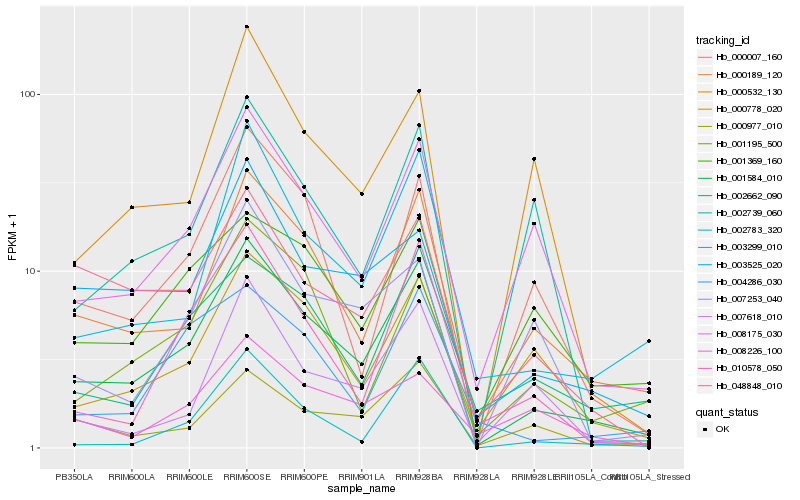
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_010 |
0.0 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 2 |
Hb_000189_120 |
0.1143203582 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 3 |
Hb_008175_030 |
0.1430757573 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 4 |
Hb_003299_010 |
0.1434803189 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 5 |
Hb_000778_020 |
0.1530531995 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 6 |
Hb_000007_160 |
0.1571412632 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 7 |
Hb_003525_020 |
0.1600427322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000977_010 |
0.1626429369 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 9 |
Hb_002662_090 |
0.16346761 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 10 |
Hb_010578_050 |
0.1649515343 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002783_320 |
0.171075755 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 12 |
Hb_001369_160 |
0.1718796294 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 13 |
Hb_001195_500 |
0.1722679918 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 14 |
Hb_048848_010 |
0.1762494992 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 15 |
Hb_004286_030 |
0.1776660614 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 16 |
Hb_007618_010 |
0.178037484 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 17 |
Hb_007253_040 |
0.1805409104 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002739_060 |
0.181411075 |
- |
- |
- |
| 19 |
Hb_000532_130 |
0.1833495482 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 20 |
Hb_008226_100 |
0.1887319762 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |