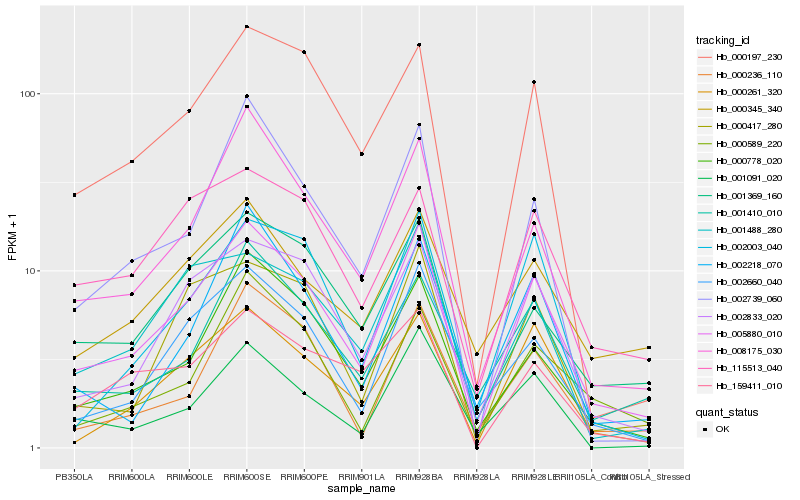
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005880_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 2 |
Hb_008175_030 |
0.1112871976 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_002833_020 |
0.1120697864 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_002660_040 |
0.1183772258 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000778_020 |
0.1199352319 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 6 |
Hb_000261_320 |
0.1252880932 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_001369_160 |
0.1271286047 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 8 |
Hb_000417_280 |
0.1332376728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000197_230 |
0.1340258081 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 10 |
Hb_002739_060 |
0.1361706122 |
- |
- |
- |
| 11 |
Hb_001410_010 |
0.1370244723 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 12 |
Hb_001488_280 |
0.1403029473 |
- |
- |
hypothetical protein JCGZ_19947 [Jatropha curcas] |
| 13 |
Hb_115513_040 |
0.142135631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000345_340 |
0.1435528058 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 15 |
Hb_001091_020 |
0.1446615551 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 16 |
Hb_159411_010 |
0.1447463115 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 17 |
Hb_002218_070 |
0.1455374732 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 18 |
Hb_000236_110 |
0.1462476434 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002003_040 |
0.1470307345 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000589_220 |
0.1477512522 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |