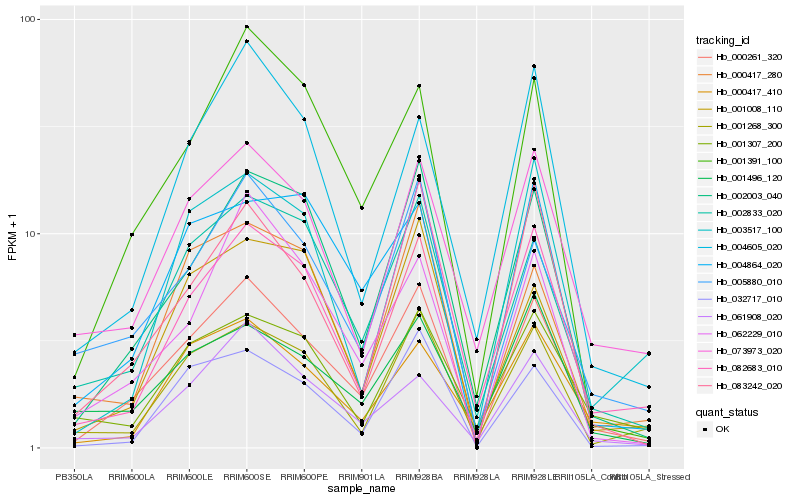
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_320 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_002003_040 |
0.1127846901 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002833_020 |
0.1170181163 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_005880_010 |
0.1252880932 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 5 |
Hb_001391_100 |
0.1288746801 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 6 |
Hb_073973_020 |
0.1380461533 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 7 |
Hb_001268_300 |
0.1381515196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_061908_020 |
0.1405655869 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_032717_010 |
0.1407751942 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_062229_010 |
0.1410930116 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 11 |
Hb_083242_020 |
0.1457400137 |
- |
- |
hypothetical protein JCGZ_20097 [Jatropha curcas] |
| 12 |
Hb_001496_120 |
0.148577715 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_082683_010 |
0.1487617383 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_004605_020 |
0.1504480328 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 15 |
Hb_000417_280 |
0.1522776501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001008_110 |
0.1524519764 |
- |
- |
- |
| 17 |
Hb_001307_200 |
0.1547578636 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 18 |
Hb_003517_100 |
0.1552728915 |
transcription factor |
TF Family: GRAS |
- |
| 19 |
Hb_000417_410 |
0.1556486133 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004864_020 |
0.1570465717 |
- |
- |
Polyprotein, putative [Solanum demissum] |