| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
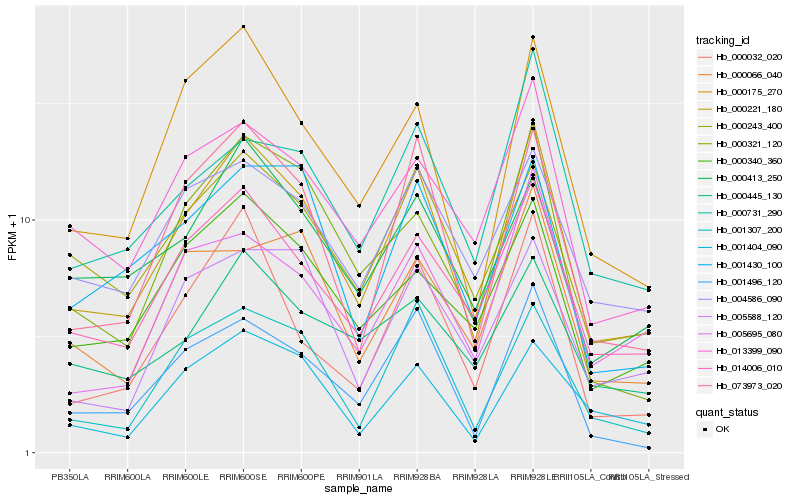
Hb_000221_180 |
0.0 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 2 |
Hb_014006_010 |
0.0901296041 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 3 |
Hb_073973_020 |
0.0902648215 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 4 |
Hb_000175_270 |
0.1026251994 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001496_120 |
0.1130370727 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_001307_200 |
0.1141078862 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 7 |
Hb_013399_090 |
0.1143186907 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_000243_400 |
0.115964651 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 9 |
Hb_000321_120 |
0.1181162164 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 10 |
Hb_000340_360 |
0.1191581426 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 11 |
Hb_000731_290 |
0.1222209472 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_004586_090 |
0.1226308673 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000032_020 |
0.1238346607 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |
| 14 |
Hb_001430_100 |
0.1250508002 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_000413_250 |
0.1262732151 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 16 |
Hb_001404_090 |
0.1264967142 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 17 |
Hb_005695_080 |
0.1265171341 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 18 |
Hb_000445_130 |
0.1301823558 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 19 |
Hb_005588_120 |
0.1311150415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000066_040 |
0.1335317055 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |