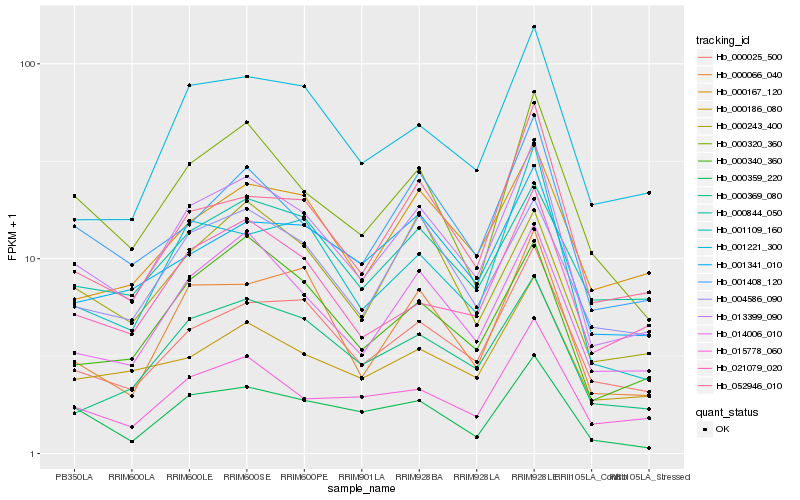
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013399_090 |
0.0 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_014006_010 |
0.0770234437 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 3 |
Hb_000340_360 |
0.0929879938 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 4 |
Hb_000025_500 |
0.0940901565 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_000320_360 |
0.0962019794 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_001408_120 |
0.0971563707 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 7 |
Hb_004586_090 |
0.0983874919 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000066_040 |
0.1015872988 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 9 |
Hb_000369_080 |
0.1018555108 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 10 |
Hb_021079_020 |
0.1061444356 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 11 |
Hb_001341_010 |
0.1068811486 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001221_300 |
0.1071412647 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_015778_060 |
0.1082712952 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 14 |
Hb_000243_400 |
0.11064784 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 15 |
Hb_000844_050 |
0.112037179 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 16 |
Hb_000359_220 |
0.1122054192 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 17 |
Hb_052946_010 |
0.1123775071 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 18 |
Hb_000167_120 |
0.1124395231 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 19 |
Hb_000186_080 |
0.11337472 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 20 |
Hb_001109_160 |
0.1137436307 |
- |
- |
DNA binding protein, putative [Ricinus communis] |