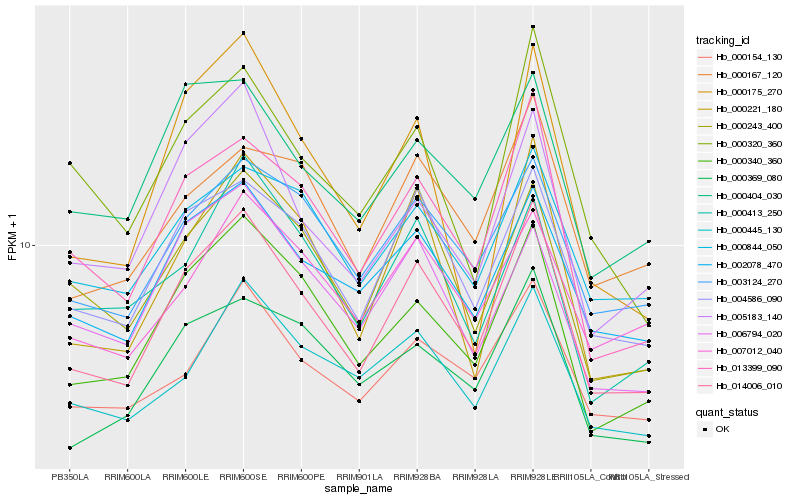
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014006_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 2 |
Hb_013399_090 |
0.0770234437 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_000340_360 |
0.0770569554 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 4 |
Hb_004586_090 |
0.0786682217 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002078_470 |
0.0896900476 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000221_180 |
0.0901296041 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 7 |
Hb_000154_130 |
0.0974845986 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 8 |
Hb_005183_140 |
0.0995000372 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 9 |
Hb_003124_270 |
0.1003457731 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 10 |
Hb_000243_400 |
0.1011743551 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 11 |
Hb_000445_130 |
0.1013532093 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 12 |
Hb_000369_080 |
0.1021534429 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 13 |
Hb_000167_120 |
0.1081933983 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 14 |
Hb_000404_030 |
0.108288001 |
- |
- |
PREDICTED: protein NAP1 [Jatropha curcas] |
| 15 |
Hb_000844_050 |
0.1095133302 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 16 |
Hb_006794_020 |
0.1106756095 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 17 |
Hb_000320_360 |
0.1128612879 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_007012_040 |
0.1137516335 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 19 |
Hb_000413_250 |
0.1138934804 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 20 |
Hb_000175_270 |
0.1151950472 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |