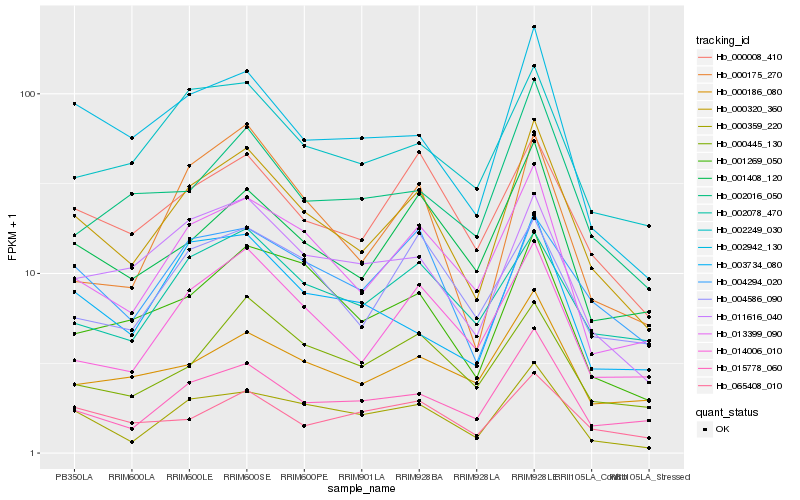
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_360 |
0.0 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_013399_090 |
0.0962019794 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_000359_220 |
0.1070487238 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 4 |
Hb_015778_060 |
0.1112166823 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 5 |
Hb_001408_120 |
0.111219442 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 6 |
Hb_002942_130 |
0.111431835 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000008_410 |
0.112675489 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_014006_010 |
0.1128612879 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 9 |
Hb_011616_040 |
0.1179226374 |
- |
- |
glucan water dikinase 3, partial [Manihot esculenta] |
| 10 |
Hb_002249_030 |
0.1220616984 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004294_020 |
0.1238527896 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001269_050 |
0.1240273676 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 13 |
Hb_002078_470 |
0.1304745663 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000186_080 |
0.1307011393 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 15 |
Hb_000445_130 |
0.1340252802 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 16 |
Hb_004586_090 |
0.1344501945 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003734_080 |
0.1350211179 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 18 |
Hb_002016_050 |
0.1350804141 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000175_270 |
0.1359546385 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 20 |
Hb_065408_010 |
0.1359574338 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |