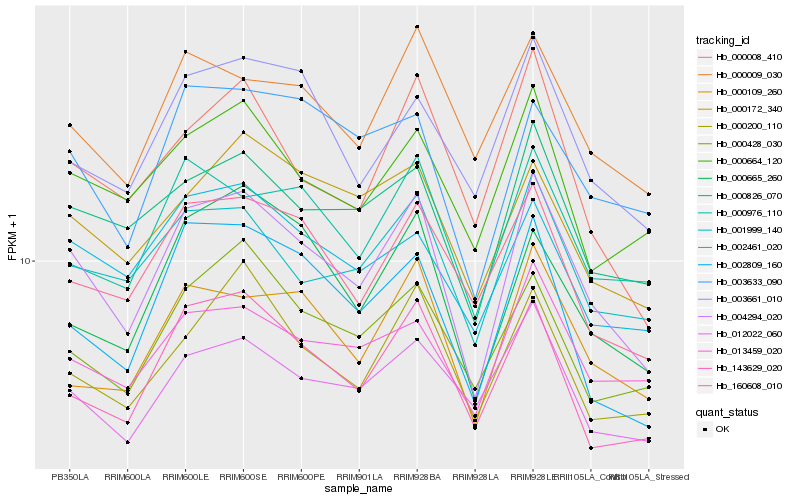
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004294_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000826_070 |
0.0932960469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_143629_020 |
0.0964654417 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 4 |
Hb_001999_140 |
0.0969570714 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_002809_160 |
0.0979773599 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 6 |
Hb_000109_260 |
0.1008843549 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_160608_010 |
0.1026728502 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 8 |
Hb_000008_410 |
0.1039379518 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_000200_110 |
0.1048133582 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000976_110 |
0.1054248926 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 11 |
Hb_000665_260 |
0.1056950826 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000664_120 |
0.1072607352 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000009_030 |
0.1074628373 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 14 |
Hb_003661_010 |
0.1082651693 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 15 |
Hb_012022_060 |
0.108719376 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003633_090 |
0.1092717866 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 17 |
Hb_000428_030 |
0.1094092302 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 18 |
Hb_000172_340 |
0.1099217361 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 19 |
Hb_013459_020 |
0.1111170704 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 20 |
Hb_002461_020 |
0.1115481315 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |