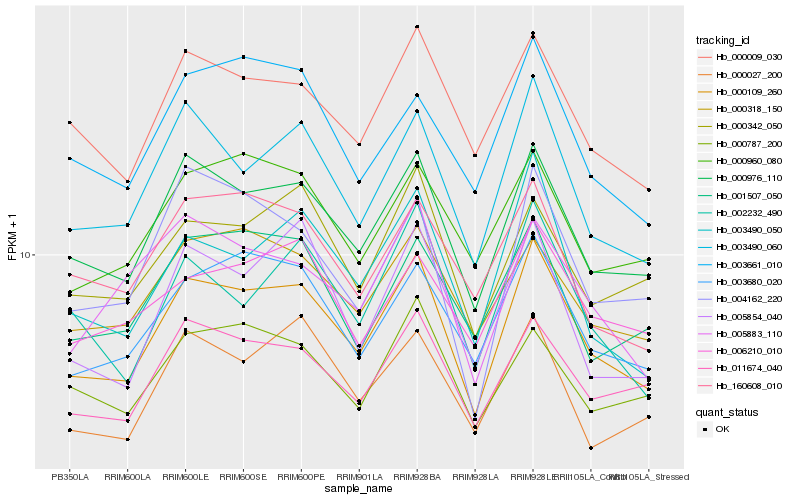
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000109_260 |
0.0 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000976_110 |
0.0551393037 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 3 |
Hb_003490_060 |
0.0698649869 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 4 |
Hb_003680_020 |
0.0782767169 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_011674_040 |
0.0819700069 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000009_030 |
0.0851615601 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 7 |
Hb_003490_050 |
0.0866576689 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005854_040 |
0.0926260591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000318_150 |
0.0938685835 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 10 |
Hb_000342_050 |
0.0948871858 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 11 |
Hb_000960_080 |
0.0954541568 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_002232_490 |
0.0957467514 |
- |
- |
PREDICTED: probable sphingolipid transporter spinster homolog 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000027_200 |
0.0958170644 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 14 |
Hb_001507_050 |
0.0979337294 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_003661_010 |
0.0982927575 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004162_220 |
0.0985566168 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_160608_010 |
0.0986209896 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 18 |
Hb_000787_200 |
0.0992030616 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_006210_010 |
0.0993649416 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 20 |
Hb_005883_110 |
0.0999535157 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |