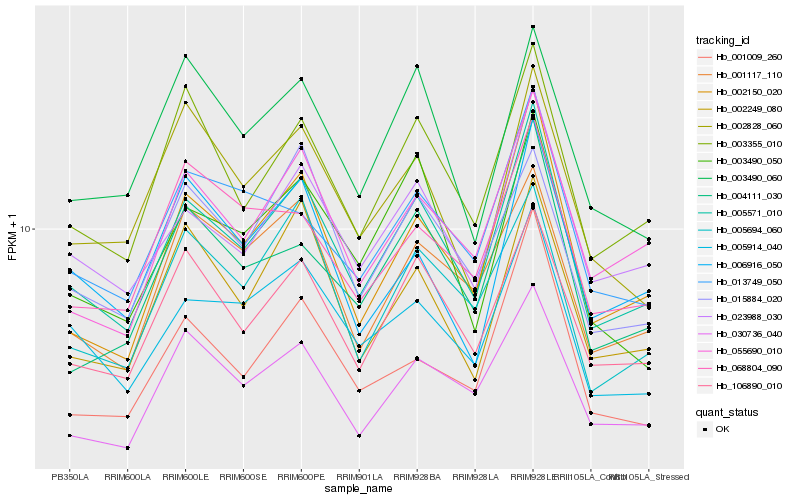
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005571_010 |
0.0 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 2 |
Hb_023988_030 |
0.0566097604 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 3 |
Hb_002150_020 |
0.0634907189 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_106890_010 |
0.0669325846 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_003355_010 |
0.0686479479 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 6 |
Hb_005914_040 |
0.0718307551 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 7 |
Hb_055690_010 |
0.0720659015 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_030736_040 |
0.0758793159 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004111_030 |
0.0824915488 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_002249_080 |
0.0826192456 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 11 |
Hb_001009_260 |
0.0849154404 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001117_110 |
0.084983123 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 13 |
Hb_005694_060 |
0.0852500336 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 14 |
Hb_013749_050 |
0.0862582683 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003490_050 |
0.0887799299 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 16 |
Hb_015884_020 |
0.0917214625 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 17 |
Hb_002828_060 |
0.0939399871 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003490_060 |
0.0945325309 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 19 |
Hb_006916_050 |
0.0953736045 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 20 |
Hb_068804_090 |
0.0956507772 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |