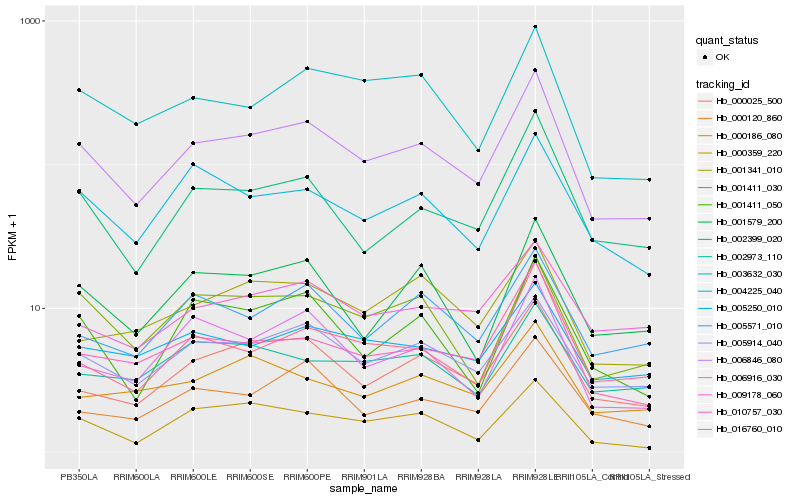
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006846_080 |
0.0 |
- |
- |
calnexin, putative [Ricinus communis] |
| 2 |
Hb_005914_040 |
0.0754932148 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 3 |
Hb_000025_500 |
0.0946652801 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 4 |
Hb_002399_020 |
0.0948525902 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 5 |
Hb_010757_030 |
0.1005265471 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_003632_030 |
0.1043814138 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 7 |
Hb_004225_040 |
0.1060774481 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 8 |
Hb_005250_010 |
0.1064840359 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 9 |
Hb_016760_010 |
0.1090151662 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_005571_010 |
0.1105647956 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 11 |
Hb_001579_200 |
0.1111231895 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 12 |
Hb_001411_030 |
0.1115338861 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 13 |
Hb_001411_050 |
0.1137941126 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 14 |
Hb_006916_030 |
0.11401864 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 15 |
Hb_009178_060 |
0.1162950158 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000120_860 |
0.1169974358 |
- |
- |
nucellin, putative [Ricinus communis] |
| 17 |
Hb_000359_220 |
0.117404446 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001341_010 |
0.1177931801 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002973_110 |
0.1198947721 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_000186_080 |
0.1199976708 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |