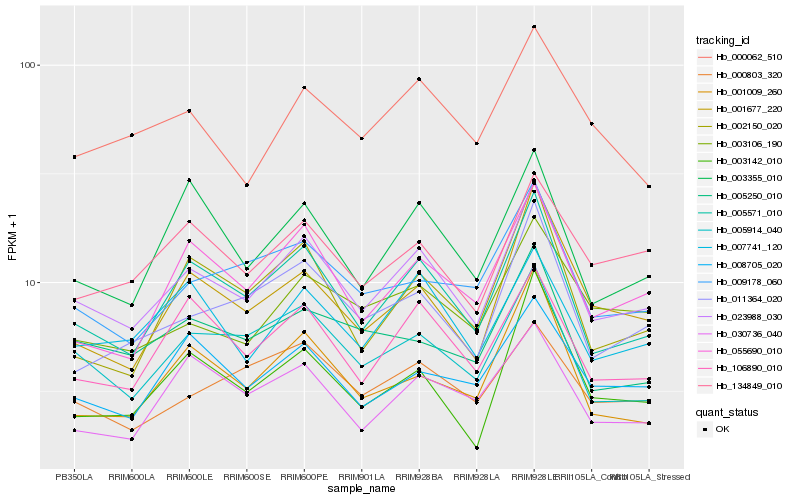
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023988_030 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 2 |
Hb_005571_010 |
0.0566097604 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 3 |
Hb_005914_040 |
0.0823658747 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 4 |
Hb_055690_010 |
0.0870045786 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_106890_010 |
0.0873318438 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_001677_220 |
0.0907129434 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 7 |
Hb_011364_020 |
0.09256116 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 8 |
Hb_007741_120 |
0.0933583597 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003355_010 |
0.0952029732 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 10 |
Hb_003106_190 |
0.0957026929 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 11 |
Hb_009178_060 |
0.0959552177 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_134849_010 |
0.0972137123 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 13 |
Hb_008705_020 |
0.0987830204 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_002150_020 |
0.100408943 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_000062_510 |
0.1017531419 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000803_320 |
0.1023944738 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 17 |
Hb_005250_010 |
0.1034144551 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 18 |
Hb_003142_010 |
0.1034828089 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 19 |
Hb_001009_260 |
0.1036857312 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_030736_040 |
0.1036879596 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |