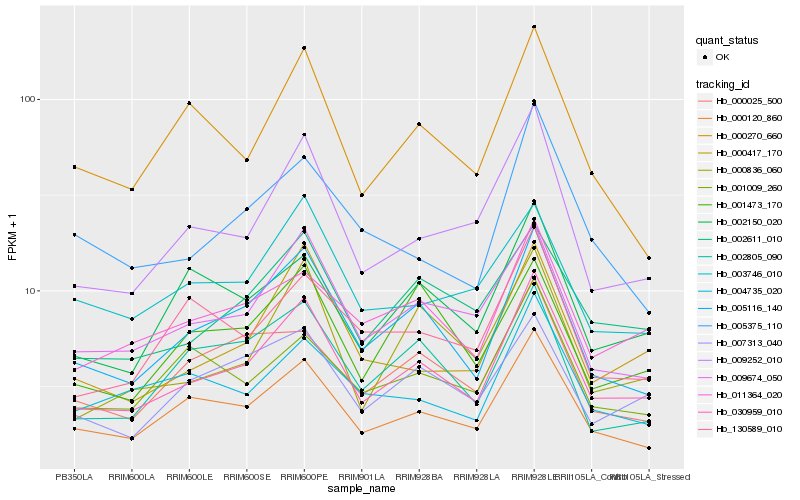
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030959_010 |
0.0 |
- |
- |
cullin, putative [Ricinus communis] |
| 2 |
Hb_005116_140 |
0.0762174917 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 3 |
Hb_000120_860 |
0.084393085 |
- |
- |
nucellin, putative [Ricinus communis] |
| 4 |
Hb_009252_010 |
0.0901115025 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 5 |
Hb_009674_050 |
0.0942229846 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 6 |
Hb_000417_170 |
0.1044115017 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |
| 7 |
Hb_002611_010 |
0.104849352 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_011364_020 |
0.1104092704 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000836_060 |
0.1126730339 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 10 |
Hb_007313_040 |
0.1165684446 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 11 |
Hb_000270_660 |
0.1197970935 |
- |
- |
purine permease, putative [Ricinus communis] |
| 12 |
Hb_002150_020 |
0.1207414682 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_002805_090 |
0.1242268809 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 14 |
Hb_004735_020 |
0.1244928915 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 15 |
Hb_001009_260 |
0.1251267795 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003746_010 |
0.1262519516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_130589_010 |
0.1270224859 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 18 |
Hb_000025_500 |
0.1275714353 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 19 |
Hb_005375_110 |
0.1287230088 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 20 |
Hb_001473_170 |
0.1289863319 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |