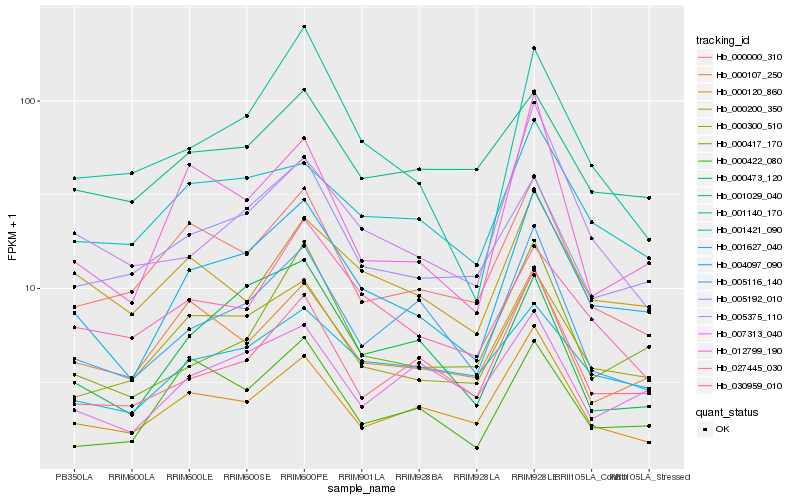
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004097_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634517 [Jatropha curcas] |
| 2 |
Hb_000300_510 |
0.1052667816 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 3 |
Hb_001627_040 |
0.1272842058 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005192_010 |
0.1279300923 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 5 |
Hb_000417_170 |
0.1292642242 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |
| 6 |
Hb_001421_090 |
0.1298480233 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 7 |
Hb_000120_860 |
0.1298588419 |
- |
- |
nucellin, putative [Ricinus communis] |
| 8 |
Hb_005116_140 |
0.1305694991 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 9 |
Hb_030959_010 |
0.1315230406 |
- |
- |
cullin, putative [Ricinus communis] |
| 10 |
Hb_012799_190 |
0.1364936339 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 11 |
Hb_000000_310 |
0.1369935614 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 12 |
Hb_001140_170 |
0.1381013444 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 13 |
Hb_000422_080 |
0.1387688006 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 14 |
Hb_007313_040 |
0.1388898386 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 15 |
Hb_000473_120 |
0.139696053 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 16 |
Hb_000107_250 |
0.1415728467 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 17 |
Hb_000200_350 |
0.1418236539 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 18 |
Hb_027445_030 |
0.1431131259 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_001029_040 |
0.1450648766 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 20 |
Hb_005375_110 |
0.1460781547 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |