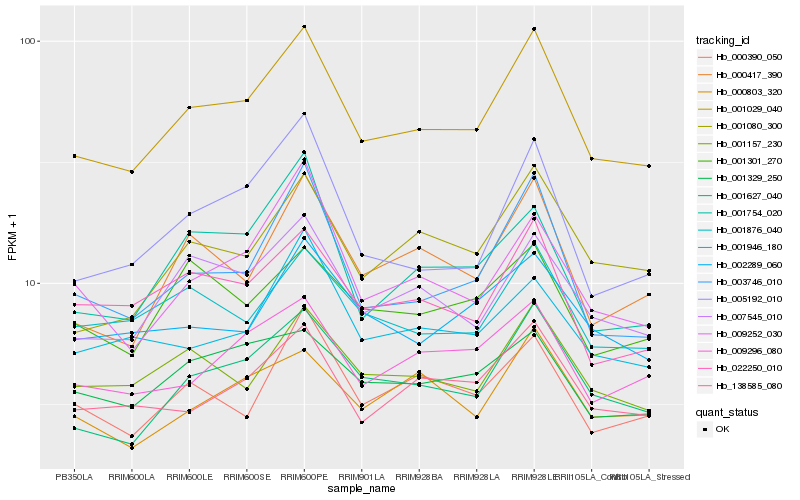
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001029_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 2 |
Hb_001627_040 |
0.062554175 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007545_010 |
0.0722108375 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 4 |
Hb_003746_010 |
0.0731207745 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001157_230 |
0.0781337249 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 6 |
Hb_138585_080 |
0.0788356119 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 7 |
Hb_022250_010 |
0.0793681435 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 8 |
Hb_000417_390 |
0.080434054 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 9 |
Hb_001080_300 |
0.0817511039 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 10 |
Hb_005192_010 |
0.0832305443 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 11 |
Hb_001876_040 |
0.0840755586 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 12 |
Hb_009296_080 |
0.0854428631 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009252_030 |
0.087916884 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 14 |
Hb_000390_050 |
0.0903803069 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 15 |
Hb_001329_250 |
0.0919851091 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002289_060 |
0.0920815463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001946_180 |
0.0933389846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000803_320 |
0.0935026325 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 19 |
Hb_001301_270 |
0.0936273576 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 20 |
Hb_001754_020 |
0.0937148232 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |