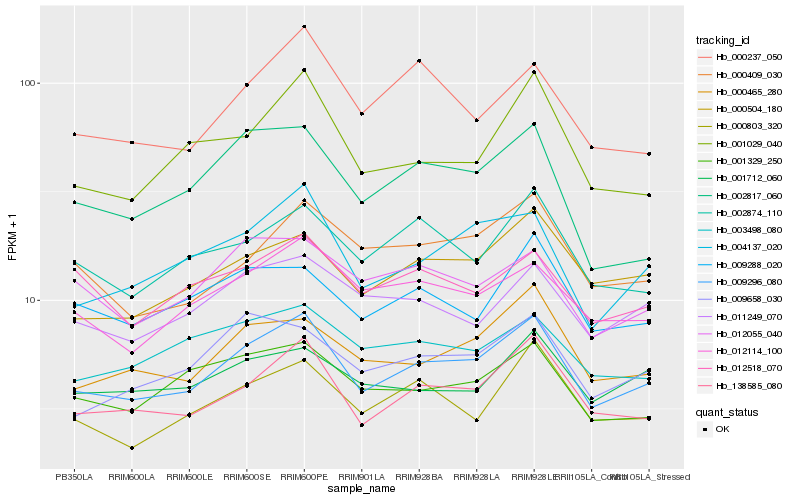
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009296_080 |
0.0 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_138585_080 |
0.0555899329 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 3 |
Hb_004137_020 |
0.0749022786 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000504_180 |
0.075046617 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 5 |
Hb_000465_280 |
0.0767036978 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 6 |
Hb_012114_100 |
0.0816848371 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 7 |
Hb_000803_320 |
0.0838717204 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 8 |
Hb_001029_040 |
0.0854428631 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 9 |
Hb_002874_110 |
0.0857252673 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 10 |
Hb_000409_030 |
0.0864418565 |
- |
- |
PREDICTED: uncharacterized protein LOC105642693 [Jatropha curcas] |
| 11 |
Hb_001329_250 |
0.0867230802 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009658_030 |
0.0877231384 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 13 |
Hb_011249_070 |
0.0890351301 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 14 |
Hb_002817_060 |
0.0899619078 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 15 |
Hb_009288_020 |
0.0900476423 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 16 |
Hb_003498_080 |
0.0921254753 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_000237_050 |
0.0932675398 |
- |
- |
CP2 [Hevea brasiliensis] |
| 18 |
Hb_012055_040 |
0.0934789061 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_001712_060 |
0.0938364009 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 20 |
Hb_012518_070 |
0.0947756449 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |