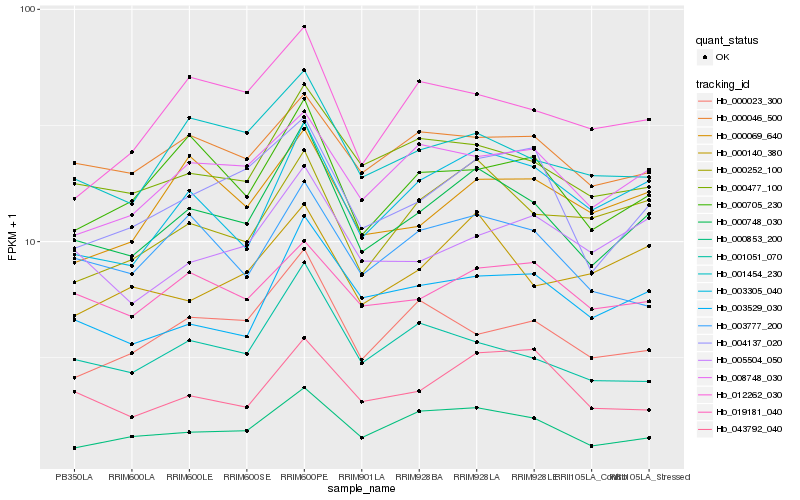
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000748_030 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000853_200 |
0.0850222351 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 3 |
Hb_000477_100 |
0.0877525733 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 4 |
Hb_001454_230 |
0.091745098 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 5 |
Hb_004137_020 |
0.0917853559 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003305_040 |
0.0937200364 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 7 |
Hb_003529_030 |
0.0947878605 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 8 |
Hb_000705_230 |
0.095244177 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 9 |
Hb_008748_030 |
0.0989670569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000046_500 |
0.1003040276 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 11 |
Hb_000252_100 |
0.1006673102 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 12 |
Hb_001051_070 |
0.1019494089 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_019181_040 |
0.1039868468 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 14 |
Hb_003777_200 |
0.1061698989 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 15 |
Hb_000069_640 |
0.1071809103 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 16 |
Hb_043792_040 |
0.1076150207 |
- |
- |
hypothetical protein, partial [Rosa rugosa] |
| 17 |
Hb_005504_050 |
0.1079575425 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 18 |
Hb_000023_300 |
0.1096191815 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 19 |
Hb_012262_030 |
0.1103867598 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000140_380 |
0.1107564464 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |