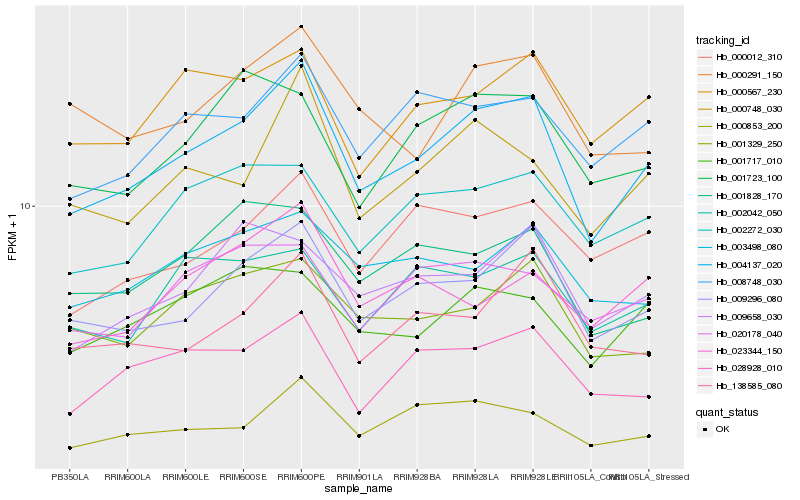
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004137_020 |
0.0 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_009296_080 |
0.0749022786 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000748_030 |
0.0917853559 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_008748_030 |
0.093909706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000853_200 |
0.0951588526 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 6 |
Hb_028928_010 |
0.0967869931 |
- |
- |
hypothetical protein CISIN_1g0030132mg, partial [Citrus sinensis] |
| 7 |
Hb_002272_030 |
0.0992216891 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 8 |
Hb_001329_250 |
0.1008864646 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000567_230 |
0.1023029767 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 10 |
Hb_000012_310 |
0.1024452926 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000291_150 |
0.1028820515 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 12 |
Hb_001717_010 |
0.1030252665 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 13 |
Hb_009658_030 |
0.1045491279 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 14 |
Hb_003498_080 |
0.1055933696 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 15 |
Hb_001723_100 |
0.1058249771 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 16 |
Hb_138585_080 |
0.10708365 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 17 |
Hb_001828_170 |
0.1072464021 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 18 |
Hb_002042_050 |
0.1075276825 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 19 |
Hb_020178_040 |
0.1075328317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_023344_150 |
0.1097355376 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |