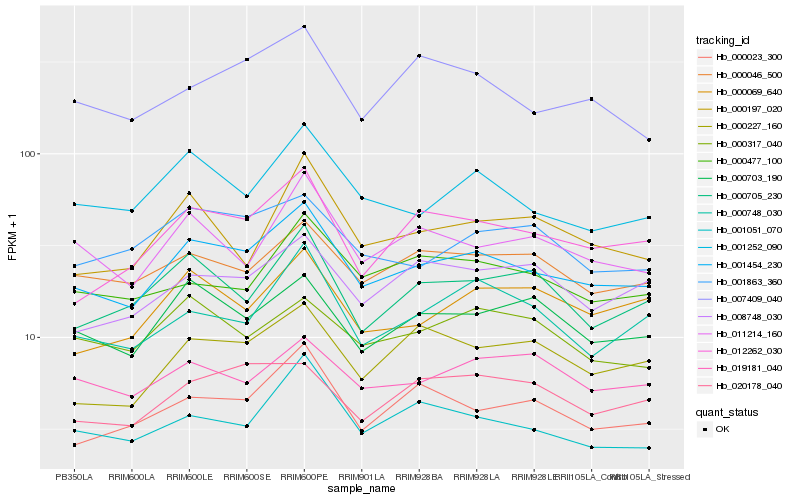
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_230 |
0.0 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 2 |
Hb_001252_090 |
0.0779260907 |
- |
- |
GDP-D-mannose pyrophosphorylase [Camellia sinensis] |
| 3 |
Hb_012262_030 |
0.0795013874 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000703_190 |
0.0819267227 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000046_500 |
0.0825923802 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 6 |
Hb_011214_160 |
0.0857884096 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 7 |
Hb_000023_300 |
0.0895190958 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 8 |
Hb_001863_360 |
0.0896220806 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 9 |
Hb_000227_160 |
0.0902644321 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 10 |
Hb_001051_070 |
0.0905577656 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000069_640 |
0.0907315243 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 12 |
Hb_000705_230 |
0.0912256819 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 13 |
Hb_000748_030 |
0.091745098 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000477_100 |
0.0924397881 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 15 |
Hb_007409_040 |
0.0929380688 |
- |
- |
actin family protein [Populus trichocarpa] |
| 16 |
Hb_008748_030 |
0.0942198034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_019181_040 |
0.0946211269 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 18 |
Hb_020178_040 |
0.096203229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000317_040 |
0.0968363768 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_000197_020 |
0.0973263401 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |