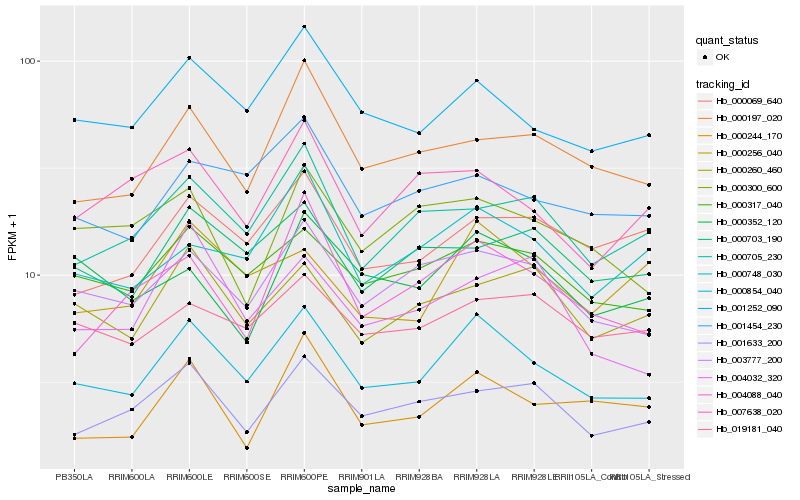
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000854_040 |
0.0 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 2 |
Hb_001252_090 |
0.0913533263 |
- |
- |
GDP-D-mannose pyrophosphorylase [Camellia sinensis] |
| 3 |
Hb_003777_200 |
0.092556329 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 4 |
Hb_000317_040 |
0.0965252052 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 5 |
Hb_001633_200 |
0.1127769577 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 6 |
Hb_001454_230 |
0.1150792598 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 7 |
Hb_000748_030 |
0.1155919265 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_000197_020 |
0.1159991149 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 9 |
Hb_000705_230 |
0.1181924014 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 10 |
Hb_004088_040 |
0.1186182757 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' iota isoform [Jatropha curcas] |
| 11 |
Hb_000256_040 |
0.1200138269 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000069_640 |
0.1207244227 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 13 |
Hb_000260_460 |
0.1216376504 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000703_190 |
0.1223086352 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_019181_040 |
0.1247897865 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 16 |
Hb_000244_170 |
0.129210635 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 17 |
Hb_004032_320 |
0.1295777876 |
- |
- |
PREDICTED: uncharacterized protein LOC105635153 [Jatropha curcas] |
| 18 |
Hb_000300_600 |
0.1300873327 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 19 |
Hb_000352_120 |
0.1301304673 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 20 |
Hb_007638_020 |
0.1302992178 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |