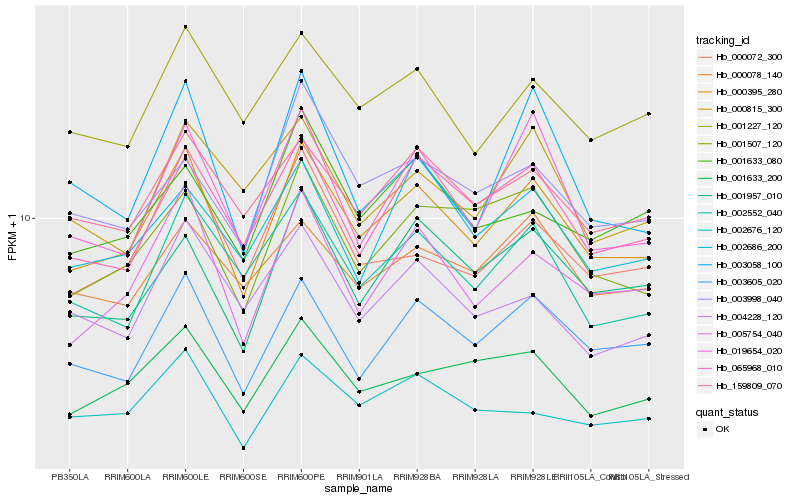
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000395_280 |
0.0 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 2 |
Hb_003605_020 |
0.0694890668 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 3 |
Hb_005754_040 |
0.0708646219 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 4 |
Hb_065968_010 |
0.0742276136 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001957_010 |
0.0801128622 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002552_040 |
0.0819516477 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 7 |
Hb_001227_120 |
0.0828812446 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 8 |
Hb_019654_020 |
0.0829175018 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 9 |
Hb_159809_070 |
0.0831200724 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 10 |
Hb_001507_120 |
0.0831798047 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_001633_200 |
0.083334229 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 12 |
Hb_000072_300 |
0.0869932288 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 13 |
Hb_002676_120 |
0.0911812927 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 14 |
Hb_000078_140 |
0.0937786049 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 15 |
Hb_003998_040 |
0.0959185491 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 16 |
Hb_001633_080 |
0.096633309 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 17 |
Hb_003058_100 |
0.0977592931 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000815_300 |
0.0980532176 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 19 |
Hb_004228_120 |
0.0985160493 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 20 |
Hb_002686_200 |
0.0992960514 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |