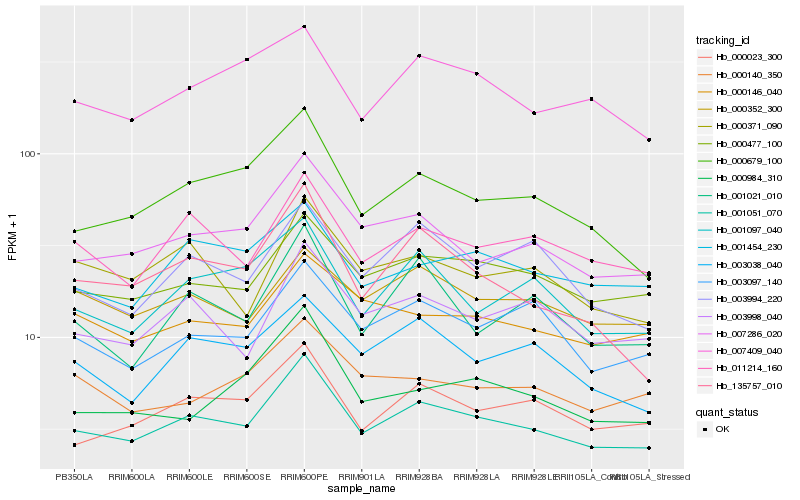
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001051_070 |
0.0 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000477_100 |
0.0725351675 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 3 |
Hb_007286_020 |
0.0731484873 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 4 |
Hb_000023_300 |
0.079294906 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 5 |
Hb_011214_160 |
0.079644704 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 6 |
Hb_003998_040 |
0.0853213567 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 7 |
Hb_000679_100 |
0.0861731135 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003097_140 |
0.0872734013 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 9 |
Hb_001454_230 |
0.0905577656 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 10 |
Hb_001097_040 |
0.0924990162 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 11 |
Hb_000146_040 |
0.0935470146 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 12 |
Hb_000352_300 |
0.0938300299 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 13 |
Hb_003038_040 |
0.0944961822 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 14 |
Hb_000371_090 |
0.0945695893 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 15 |
Hb_003994_220 |
0.0962004328 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 16 |
Hb_007409_040 |
0.0962760553 |
- |
- |
actin family protein [Populus trichocarpa] |
| 17 |
Hb_001021_010 |
0.0966724247 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 18 |
Hb_135757_010 |
0.0990857227 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 19 |
Hb_000140_350 |
0.099797389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000984_310 |
0.1001128637 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |