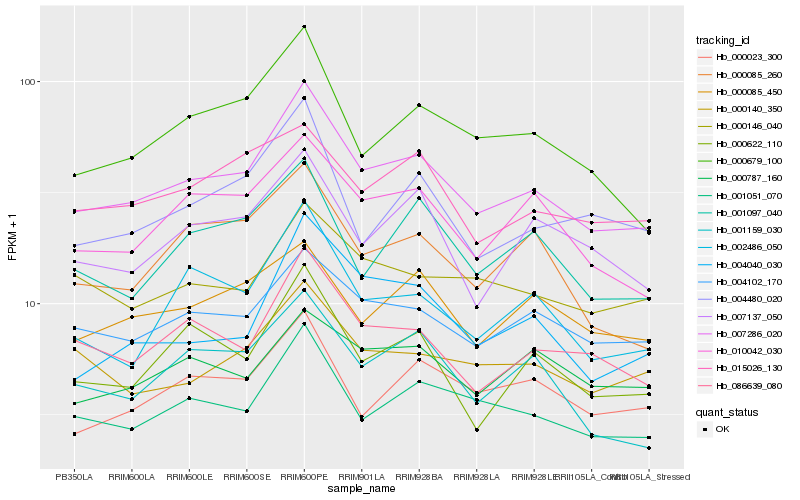
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007286_020 |
0.0 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 2 |
Hb_001051_070 |
0.0731484873 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004102_170 |
0.0759863142 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 4 |
Hb_086639_080 |
0.0786722709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010042_030 |
0.079590372 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 6 |
Hb_000679_100 |
0.0852669209 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002486_050 |
0.0879164009 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000622_110 |
0.0884985021 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 9 |
Hb_007137_050 |
0.0905630186 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_000085_450 |
0.0908981478 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 11 |
Hb_000023_300 |
0.0911131118 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 12 |
Hb_001097_040 |
0.0921959476 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 13 |
Hb_015026_130 |
0.0932260003 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 14 |
Hb_004040_030 |
0.0935171832 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000140_350 |
0.093796659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004480_020 |
0.0941869503 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 17 |
Hb_000146_040 |
0.0942672555 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 18 |
Hb_001159_030 |
0.0944269919 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 19 |
Hb_000085_260 |
0.095878528 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 20 |
Hb_000787_160 |
0.0966183089 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |