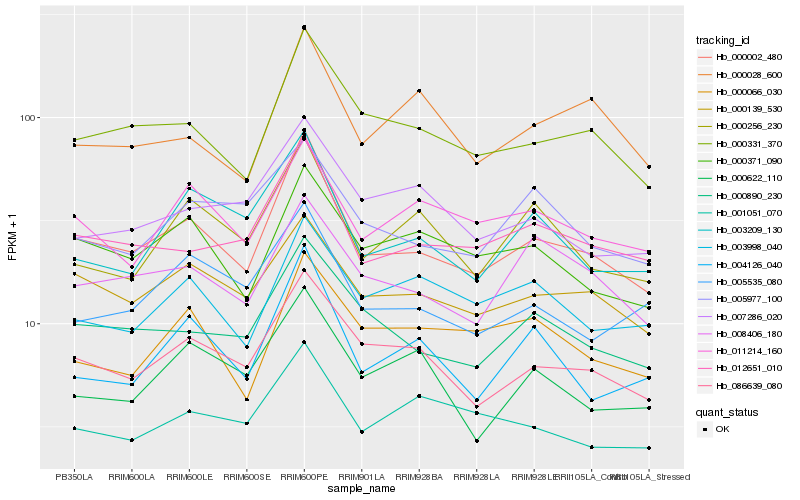
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_480 |
0.0 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000331_370 |
0.0747892493 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 3 |
Hb_086639_080 |
0.0801494652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012651_010 |
0.0851836222 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 5 |
Hb_000890_230 |
0.0875865291 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 6 |
Hb_000139_530 |
0.0918381497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000371_090 |
0.099606184 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 8 |
Hb_004126_040 |
0.1014692258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003209_130 |
0.1039522575 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 10 |
Hb_008406_180 |
0.104495379 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 11 |
Hb_000256_230 |
0.1084335086 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 12 |
Hb_005535_080 |
0.1092740853 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 13 |
Hb_011214_160 |
0.1096258945 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 14 |
Hb_003998_040 |
0.1129950501 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 15 |
Hb_001051_070 |
0.1132787219 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007286_020 |
0.1155229782 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 17 |
Hb_000028_600 |
0.1163719037 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 18 |
Hb_000622_110 |
0.1166536313 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 19 |
Hb_000066_030 |
0.1173560652 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005977_100 |
0.1175861599 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |