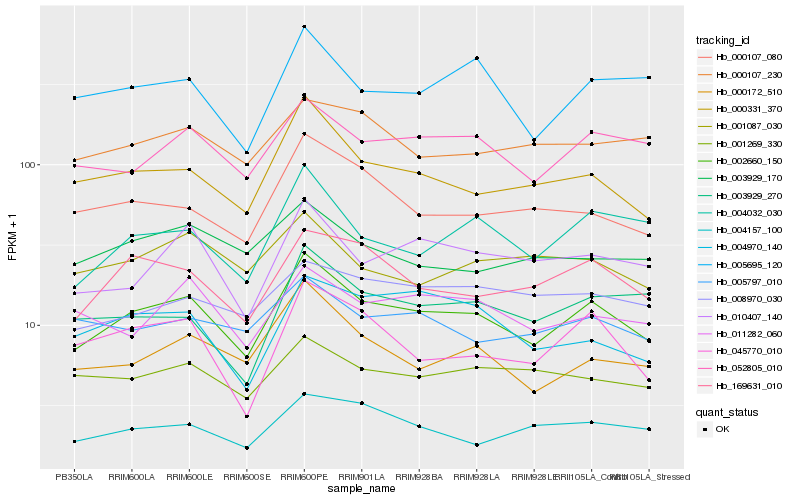
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002660_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000331_370 |
0.0929483335 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 3 |
Hb_052805_010 |
0.0954282497 |
- |
- |
hypothetical protein POPTR_0009s03650g [Populus trichocarpa] |
| 4 |
Hb_169631_010 |
0.0975819877 |
- |
- |
MADS-box transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000172_510 |
0.0999463386 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |
| 6 |
Hb_004032_030 |
0.1033884069 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 7 |
Hb_010407_140 |
0.1060326695 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 8 |
Hb_000107_080 |
0.1082092022 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 9 |
Hb_045770_010 |
0.1098319221 |
- |
- |
ER lumen protein retaining receptor [Populus trichocarpa] |
| 10 |
Hb_005695_120 |
0.1119188525 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 11 |
Hb_011282_060 |
0.1152987523 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 12 |
Hb_003929_270 |
0.1164366102 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001269_330 |
0.1169780809 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 14 |
Hb_004157_100 |
0.1177559028 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 15 |
Hb_005797_010 |
0.1191928115 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_004970_140 |
0.1196340151 |
- |
- |
PREDICTED: uncharacterized protein LOC105633635 [Jatropha curcas] |
| 17 |
Hb_008970_030 |
0.1210577987 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 18 |
Hb_001087_030 |
0.1218824842 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 19 |
Hb_003929_170 |
0.1224617006 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 20 |
Hb_000107_230 |
0.1225537211 |
- |
- |
unknown [Medicago truncatula] |