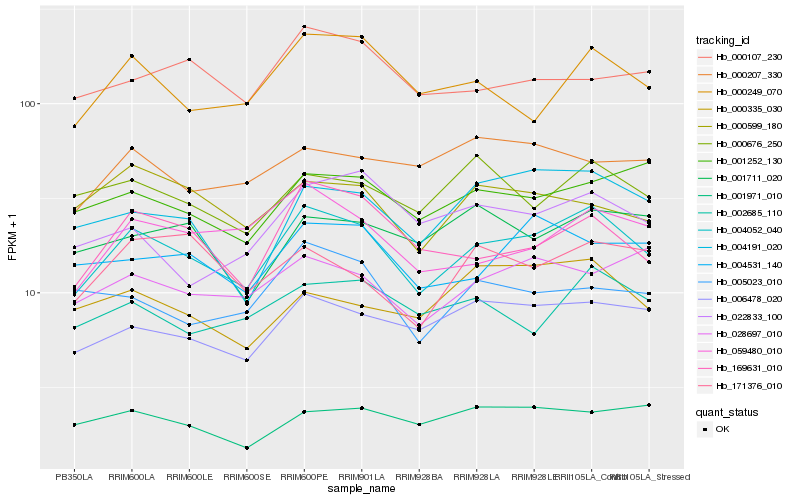
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004052_040 |
0.0 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 2 |
Hb_006478_020 |
0.0988455784 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 3 |
Hb_169631_010 |
0.102574 |
- |
- |
MADS-box transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000249_070 |
0.1063449652 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 5 |
Hb_059480_010 |
0.1077719884 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 6 |
Hb_004191_020 |
0.1097946749 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 7 |
Hb_002685_110 |
0.1133737535 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 8 |
Hb_001252_130 |
0.1156176655 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 9 |
Hb_004531_140 |
0.1168194989 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 10 |
Hb_000676_250 |
0.1173876847 |
- |
- |
PREDICTED: protein RTE1-HOMOLOG isoform X1 [Jatropha curcas] |
| 11 |
Hb_028697_010 |
0.1191111282 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 12 |
Hb_005023_010 |
0.1219798422 |
- |
- |
PREDICTED: putative lipase ROG1 [Jatropha curcas] |
| 13 |
Hb_000599_180 |
0.1230006141 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_000107_230 |
0.1232155882 |
- |
- |
unknown [Medicago truncatula] |
| 15 |
Hb_001971_010 |
0.1241580488 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 16 |
Hb_171376_010 |
0.1249082608 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 17 |
Hb_022833_100 |
0.1250153423 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_000335_030 |
0.1250829617 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17033 [Jatropha curcas] |
| 19 |
Hb_001711_020 |
0.1259975528 |
- |
- |
PREDICTED: AP-2 complex subunit mu [Jatropha curcas] |
| 20 |
Hb_000207_330 |
0.126632688 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |