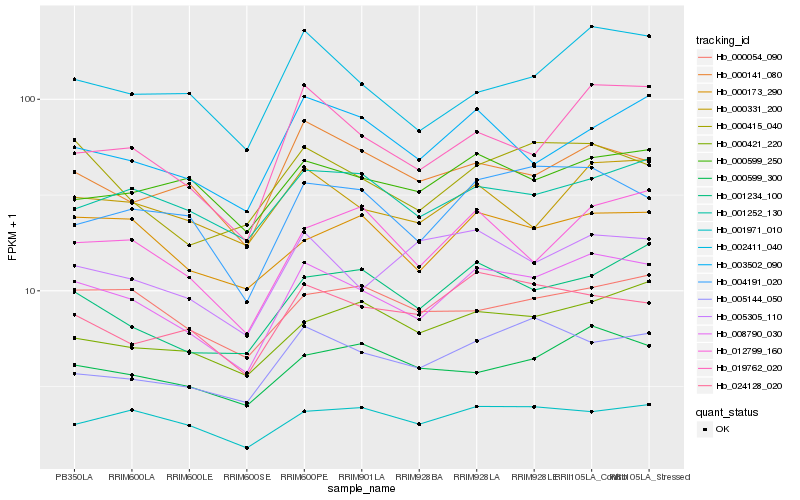
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008790_030 |
0.0 |
- |
- |
PREDICTED: probable magnesium transporter NIPA9 [Jatropha curcas] |
| 2 |
Hb_000415_040 |
0.0908318417 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 3 |
Hb_004191_020 |
0.0918807104 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 4 |
Hb_000331_200 |
0.0941502497 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 5 |
Hb_019762_020 |
0.0955835005 |
- |
- |
PREDICTED: selenoprotein K-like [Nicotiana tomentosiformis] |
| 6 |
Hb_000173_290 |
0.096701942 |
- |
- |
guanylate kinase, putative [Ricinus communis] |
| 7 |
Hb_024128_020 |
0.0976102823 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002411_040 |
0.0979813148 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 6 [Jatropha curcas] |
| 9 |
Hb_001234_100 |
0.0982047982 |
- |
- |
PREDICTED: eyes absent homolog 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000599_300 |
0.0997781071 |
- |
- |
PREDICTED: TBC1 domain family member 15-like [Jatropha curcas] |
| 11 |
Hb_005144_050 |
0.0999685273 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |
| 12 |
Hb_000141_080 |
0.1009367578 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial isoform X1 [Brassica rapa] |
| 13 |
Hb_012799_160 |
0.1012704171 |
- |
- |
PREDICTED: ADP-ribosylation factor-related protein 1-like [Jatropha curcas] |
| 14 |
Hb_001971_010 |
0.1026303946 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 15 |
Hb_005305_110 |
0.1027015748 |
- |
- |
PREDICTED: uncharacterized protein LOC105648286 [Jatropha curcas] |
| 16 |
Hb_003502_090 |
0.1030406376 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 17 |
Hb_001252_130 |
0.1032737557 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 18 |
Hb_000421_220 |
0.1041990367 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000054_090 |
0.104312831 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000599_250 |
0.1046383666 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |