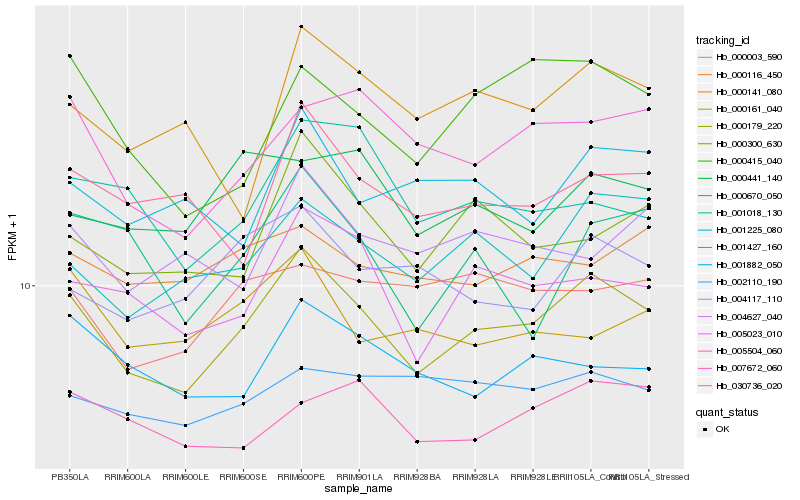
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000179_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized membrane protein C776.05 [Jatropha curcas] |
| 2 |
Hb_005023_010 |
0.0979100874 |
- |
- |
PREDICTED: putative lipase ROG1 [Jatropha curcas] |
| 3 |
Hb_001225_080 |
0.1001475212 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8A [Populus euphratica] |
| 4 |
Hb_005504_060 |
0.108306404 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Populus euphratica] |
| 5 |
Hb_000415_040 |
0.10928162 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 6 |
Hb_000300_630 |
0.111631 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 7 |
Hb_004117_110 |
0.1137263251 |
- |
- |
hypothetical protein POPTR_0016s14420g [Populus trichocarpa] |
| 8 |
Hb_030736_020 |
0.1144297934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001882_050 |
0.1156172808 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000003_590 |
0.1173632079 |
- |
- |
PREDICTED: uncharacterized protein LOC105631550 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007672_060 |
0.1180908561 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
Upstream activation factor subunit UAF30, putative [Ricinus communis] |
| 12 |
Hb_001427_160 |
0.118704304 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000141_080 |
0.1196340244 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial isoform X1 [Brassica rapa] |
| 14 |
Hb_000670_050 |
0.1196819846 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 15 |
Hb_004627_040 |
0.1201840853 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 16 |
Hb_000116_450 |
0.1206125491 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_002110_190 |
0.1207113846 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27460 [Jatropha curcas] |
| 18 |
Hb_000441_140 |
0.1213115315 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-7-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001018_130 |
0.1218336897 |
- |
- |
PREDICTED: uncharacterized protein LOC105638260 [Jatropha curcas] |
| 20 |
Hb_000161_040 |
0.1229279972 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |