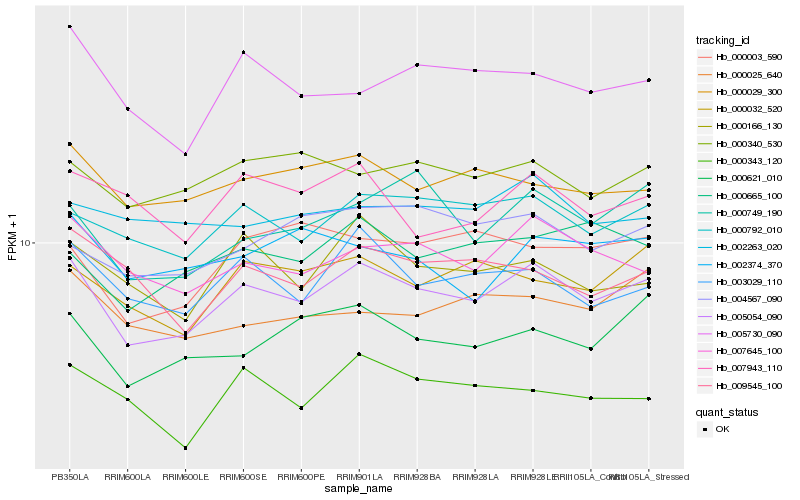
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005054_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At2g39910 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000792_010 |
0.0662097506 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 3 |
Hb_007645_100 |
0.0702750643 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003029_110 |
0.0716007807 |
- |
- |
PREDICTED: uncharacterized protein LOC105640062 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000621_010 |
0.0761074484 |
- |
- |
PREDICTED: uncharacterized protein LOC103444129 [Malus domestica] |
| 6 |
Hb_000029_300 |
0.0765105024 |
- |
- |
hypothetical protein JCGZ_10048 [Jatropha curcas] |
| 7 |
Hb_000032_520 |
0.0769070815 |
- |
- |
PREDICTED: F-box protein SKIP8-like [Jatropha curcas] |
| 8 |
Hb_000665_100 |
0.0786923258 |
- |
- |
PREDICTED: neurochondrin [Jatropha curcas] |
| 9 |
Hb_000340_530 |
0.0792555414 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |
| 10 |
Hb_000749_190 |
0.0792660112 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Populus euphratica] |
| 11 |
Hb_000166_130 |
0.0800924044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005730_090 |
0.0815126333 |
- |
- |
PREDICTED: uncharacterized protein LOC105631484 [Jatropha curcas] |
| 13 |
Hb_004567_090 |
0.0831775495 |
- |
- |
PREDICTED: GPI transamidase component PIG-T [Jatropha curcas] |
| 14 |
Hb_009545_100 |
0.0834860634 |
- |
- |
PREDICTED: protein OBERON 1 [Jatropha curcas] |
| 15 |
Hb_000003_590 |
0.0841050382 |
- |
- |
PREDICTED: uncharacterized protein LOC105631550 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007943_110 |
0.0853731048 |
- |
- |
PREDICTED: basic salivary proline-rich protein 3 [Jatropha curcas] |
| 17 |
Hb_000025_640 |
0.0854563684 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002374_370 |
0.0861319193 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000343_120 |
0.0867781806 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16010 [Jatropha curcas] |
| 20 |
Hb_002263_020 |
0.0873076742 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |