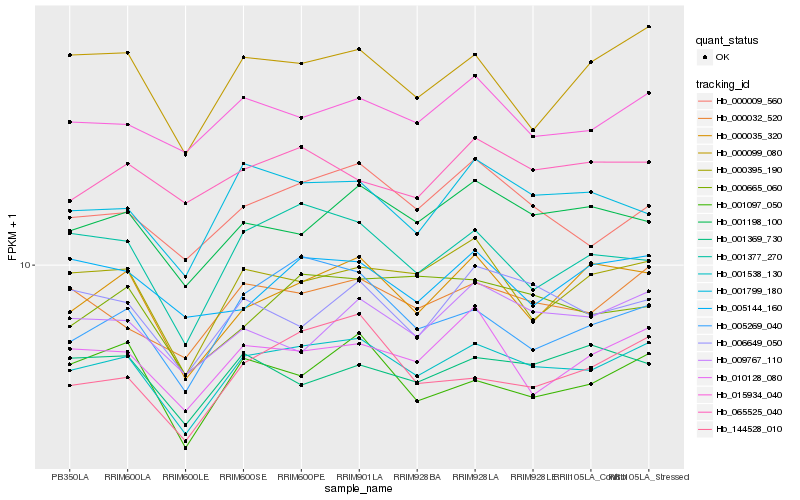
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001538_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000032_520 |
0.0597714737 |
- |
- |
PREDICTED: F-box protein SKIP8-like [Jatropha curcas] |
| 3 |
Hb_001198_100 |
0.0661698264 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7-like [Jatropha curcas] |
| 4 |
Hb_000395_190 |
0.0670057714 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 7-like [Jatropha curcas] |
| 5 |
Hb_000009_560 |
0.0677520605 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 6 |
Hb_015934_040 |
0.067827519 |
- |
- |
PREDICTED: protein Hook homolog 3-like [Jatropha curcas] |
| 7 |
Hb_000035_320 |
0.0699449905 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22 [Jatropha curcas] |
| 8 |
Hb_001799_180 |
0.0701878862 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 9 |
Hb_000099_080 |
0.0707596463 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 10 |
Hb_010128_080 |
0.0727431741 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001377_270 |
0.0728051641 |
- |
- |
hypothetical protein CICLE_v10021159mg [Citrus clementina] |
| 12 |
Hb_001097_050 |
0.0751395487 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_006649_050 |
0.0757119547 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 14 |
Hb_000665_060 |
0.0766109866 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 15 |
Hb_144528_010 |
0.0766143594 |
- |
- |
type II inositol 5-phosphatase, putative [Ricinus communis] |
| 16 |
Hb_009767_110 |
0.0776098245 |
- |
- |
- |
| 17 |
Hb_005269_040 |
0.0786407109 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein At1g06270 [Jatropha curcas] |
| 18 |
Hb_065525_040 |
0.079677125 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 19 |
Hb_001369_730 |
0.0800364718 |
- |
- |
F5O11.10, putative isoform 1 [Theobroma cacao] |
| 20 |
Hb_005144_160 |
0.0812115673 |
- |
- |
PREDICTED: uncharacterized protein LOC105628018 isoform X1 [Jatropha curcas] |