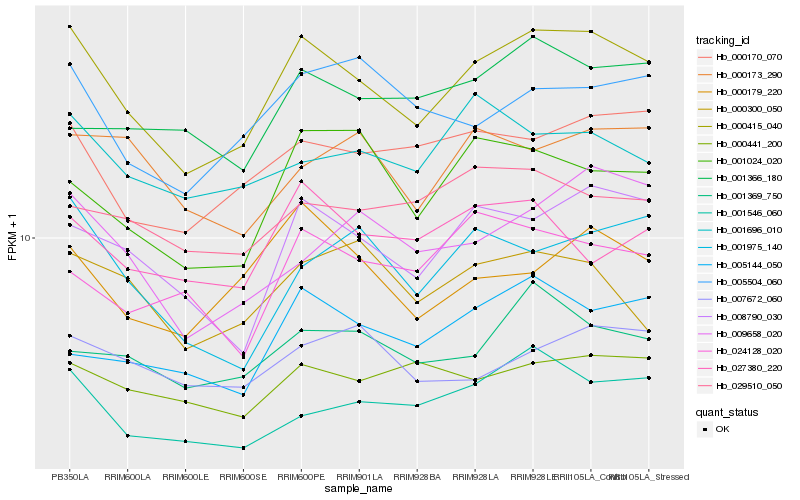
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000415_040 |
0.0 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 2 |
Hb_008790_030 |
0.0908318417 |
- |
- |
PREDICTED: probable magnesium transporter NIPA9 [Jatropha curcas] |
| 3 |
Hb_000170_070 |
0.1011414334 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At4g08455 [Jatropha curcas] |
| 4 |
Hb_001546_060 |
0.1089388663 |
- |
- |
PREDICTED: uncharacterized protein LOC105133943 isoform X4 [Populus euphratica] |
| 5 |
Hb_000179_220 |
0.10928162 |
- |
- |
PREDICTED: uncharacterized membrane protein C776.05 [Jatropha curcas] |
| 6 |
Hb_001369_750 |
0.110444079 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 7 |
Hb_005504_060 |
0.1113069971 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Populus euphratica] |
| 8 |
Hb_000441_200 |
0.1123050609 |
- |
- |
PREDICTED: small RNA 2'-O-methyltransferase-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001024_020 |
0.1136770054 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 10 |
Hb_001975_140 |
0.1149951332 |
- |
- |
PREDICTED: urease accessory protein F [Jatropha curcas] |
| 11 |
Hb_005144_050 |
0.1158975309 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |
| 12 |
Hb_027380_220 |
0.1167836927 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 13 |
Hb_001696_010 |
0.1168096205 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_009658_020 |
0.1168640156 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15 [Jatropha curcas] |
| 15 |
Hb_007672_060 |
0.1174357571 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
Upstream activation factor subunit UAF30, putative [Ricinus communis] |
| 16 |
Hb_029510_050 |
0.1187183239 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 17 |
Hb_000300_050 |
0.1188173352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000173_290 |
0.1200989106 |
- |
- |
guanylate kinase, putative [Ricinus communis] |
| 19 |
Hb_024128_020 |
0.1228880713 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001366_180 |
0.1245399967 |
- |
- |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |