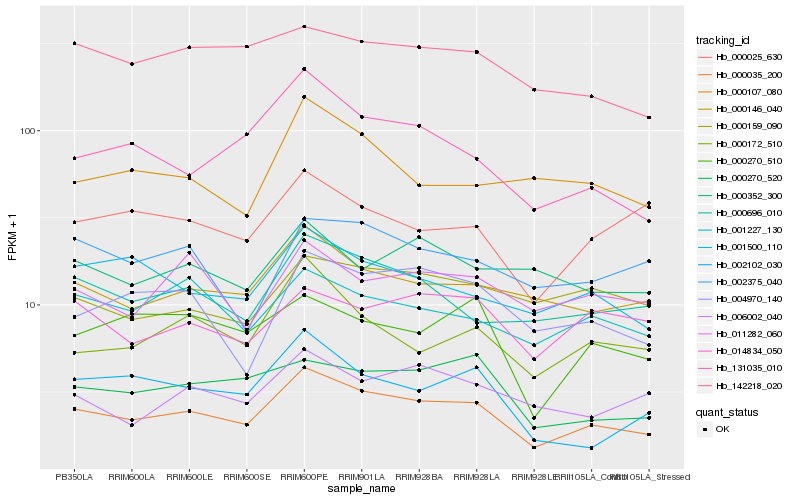
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_200 |
0.0 |
- |
- |
PREDICTED: phospholipase D Z-like [Jatropha curcas] |
| 2 |
Hb_000146_040 |
0.101501233 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 3 |
Hb_000696_010 |
0.102886103 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 4 |
Hb_131035_010 |
0.1098427943 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_001500_110 |
0.1115318883 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000270_520 |
0.1133395911 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004970_140 |
0.1139689619 |
- |
- |
PREDICTED: uncharacterized protein LOC105633635 [Jatropha curcas] |
| 8 |
Hb_000172_510 |
0.1147437882 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |
| 9 |
Hb_001227_130 |
0.1155459804 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 10 |
Hb_142218_020 |
0.1190788656 |
- |
- |
actin family protein [Populus trichocarpa] |
| 11 |
Hb_006002_040 |
0.1207691009 |
- |
- |
PREDICTED: uncharacterized protein LOC105638085 [Jatropha curcas] |
| 12 |
Hb_000270_510 |
0.1235339329 |
- |
- |
hypothetical protein CISIN_1g008173mg [Citrus sinensis] |
| 13 |
Hb_011282_060 |
0.1236615162 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 14 |
Hb_014834_050 |
0.1246892849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002102_030 |
0.1256131488 |
- |
- |
- |
| 16 |
Hb_000159_090 |
0.1256347128 |
- |
- |
hypothetical protein POPTR_0015s153201g, partial [Populus trichocarpa] |
| 17 |
Hb_000107_080 |
0.1269233587 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 18 |
Hb_002375_040 |
0.1270463478 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 19 |
Hb_000025_630 |
0.1283257181 |
- |
- |
PREDICTED: uncharacterized protein LOC105635952 [Jatropha curcas] |
| 20 |
Hb_000352_300 |
0.1284977308 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |