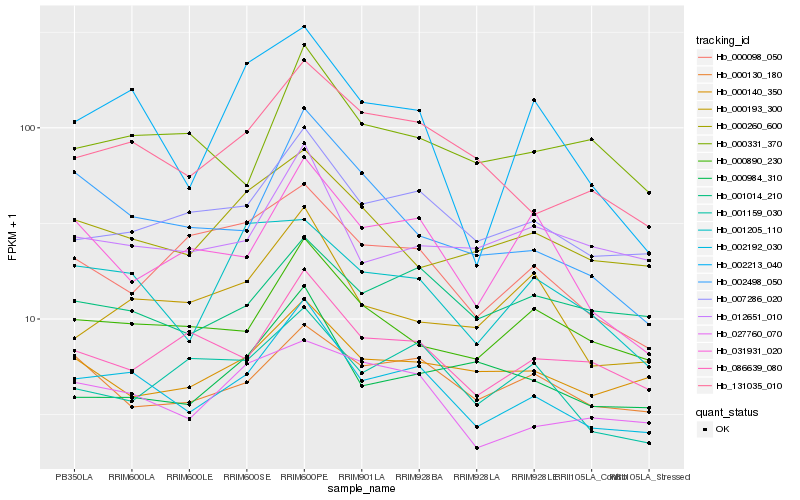
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002192_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007286_020 |
0.1053674975 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 3 |
Hb_131035_010 |
0.1102982009 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000890_230 |
0.1236353404 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 5 |
Hb_086639_080 |
0.1287355156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_027760_070 |
0.13045741 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 7 |
Hb_000193_300 |
0.1343467472 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 8 |
Hb_012651_010 |
0.1350140893 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 9 |
Hb_000260_600 |
0.1366886048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002498_050 |
0.1371615855 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 11 |
Hb_000130_180 |
0.1374804145 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000140_350 |
0.1387367369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001014_210 |
0.1388919325 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 14 |
Hb_031931_020 |
0.1391151573 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 15 |
Hb_001205_110 |
0.1399691437 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 16 |
Hb_000984_310 |
0.1399937378 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 17 |
Hb_000098_050 |
0.1401880308 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 18 |
Hb_002213_040 |
0.1401924273 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 19 |
Hb_000331_370 |
0.1410231439 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 20 |
Hb_001159_030 |
0.1413913672 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |