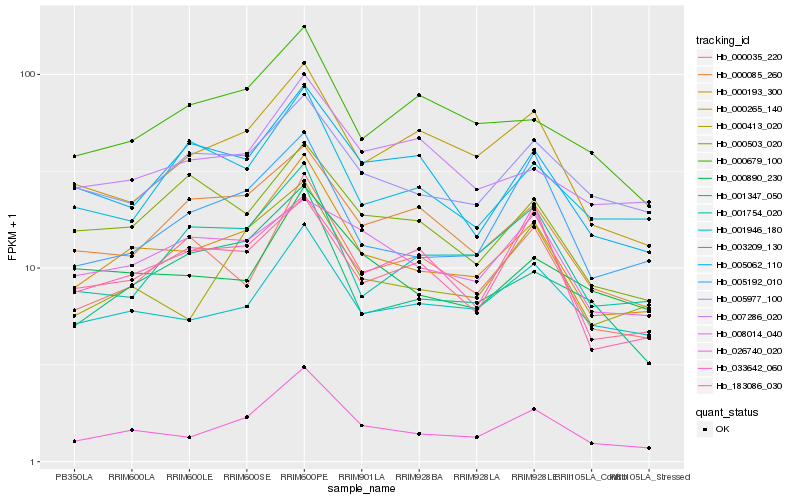
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000193_300 |
0.0 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 2 |
Hb_026740_020 |
0.0745132877 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_000413_020 |
0.1015437476 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 4 |
Hb_001946_180 |
0.1017497468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005977_100 |
0.1054337458 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 6 |
Hb_001347_050 |
0.1062136174 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000085_260 |
0.1066486026 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 8 |
Hb_000035_220 |
0.1068045226 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 9 |
Hb_005192_010 |
0.1092720772 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_000265_140 |
0.1105887933 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 11 |
Hb_000890_230 |
0.1128131395 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 12 |
Hb_183086_030 |
0.1135007329 |
- |
- |
GTPase-activating protein GYP7 [Gossypium arboreum] |
| 13 |
Hb_007286_020 |
0.1148587561 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 14 |
Hb_000679_100 |
0.115252917 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005062_110 |
0.1153798277 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 16 |
Hb_003209_130 |
0.1158756632 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 17 |
Hb_001754_020 |
0.1160966147 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 18 |
Hb_000503_020 |
0.1165395608 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 19 |
Hb_033642_060 |
0.1194699155 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_008014_040 |
0.1206617267 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |