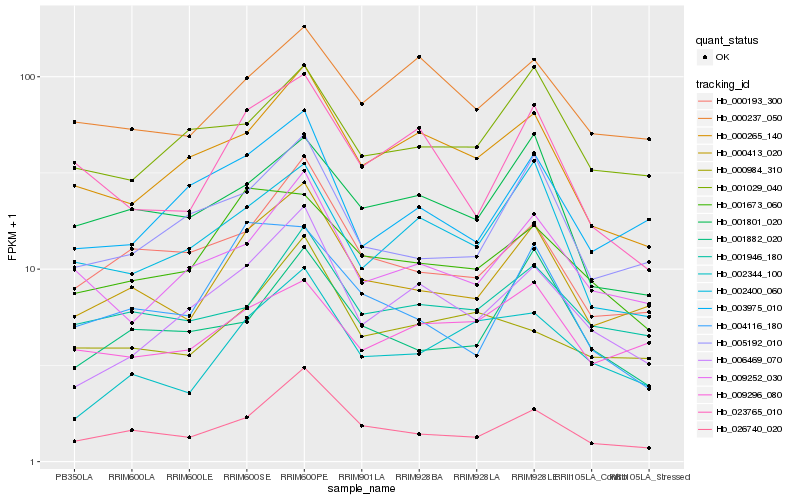
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000413_020 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 2 |
Hb_026740_020 |
0.0901444931 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_000193_300 |
0.1015437476 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 4 |
Hb_005192_010 |
0.1032513134 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 5 |
Hb_003975_010 |
0.11671567 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 6 |
Hb_001946_180 |
0.1173870247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009252_030 |
0.120120874 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 8 |
Hb_001673_060 |
0.1294196252 |
- |
- |
PREDICTED: uncharacterized protein LOC105635789 [Jatropha curcas] |
| 9 |
Hb_002344_100 |
0.1311397421 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 10 |
Hb_000265_140 |
0.1322687779 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 11 |
Hb_009296_080 |
0.1334902277 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001801_020 |
0.1342256072 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_001029_040 |
0.1350721941 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 14 |
Hb_001882_020 |
0.1354229761 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 15 |
Hb_006469_070 |
0.1355521121 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 16 |
Hb_004116_180 |
0.1357292158 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 17 |
Hb_023765_010 |
0.1365717831 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 18 |
Hb_000237_050 |
0.1368799092 |
- |
- |
CP2 [Hevea brasiliensis] |
| 19 |
Hb_002400_060 |
0.1369046654 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 20 |
Hb_000984_310 |
0.1377429322 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |