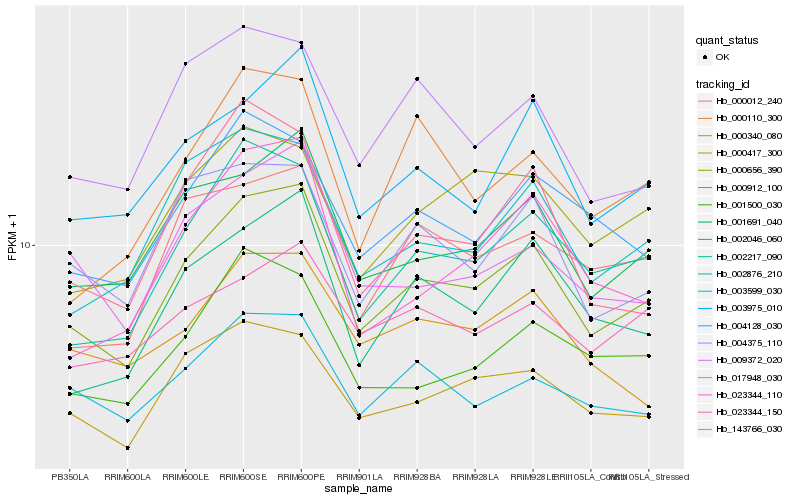
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000912_100 |
0.0 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 2 |
Hb_001691_040 |
0.081253803 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002217_090 |
0.0832357797 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 4 |
Hb_000417_300 |
0.0873319245 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 [Jatropha curcas] |
| 5 |
Hb_003975_010 |
0.0906771909 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 6 |
Hb_002046_060 |
0.0960390254 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 7 |
Hb_002876_210 |
0.0962458745 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 8 |
Hb_003599_030 |
0.0977791304 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_004128_030 |
0.0983042394 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004375_110 |
0.1017586803 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 11 |
Hb_017948_030 |
0.102065344 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 12 |
Hb_000012_240 |
0.1033508674 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_009372_020 |
0.1056035245 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 14 |
Hb_001500_030 |
0.1068409216 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 15 |
Hb_023344_110 |
0.1085286618 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 16 |
Hb_000656_390 |
0.1104408334 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000340_080 |
0.1126471267 |
- |
- |
PREDICTED: isoamylase 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_143766_030 |
0.1135207369 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000110_300 |
0.1137601338 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 20 |
Hb_023344_150 |
0.1141613526 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |