| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
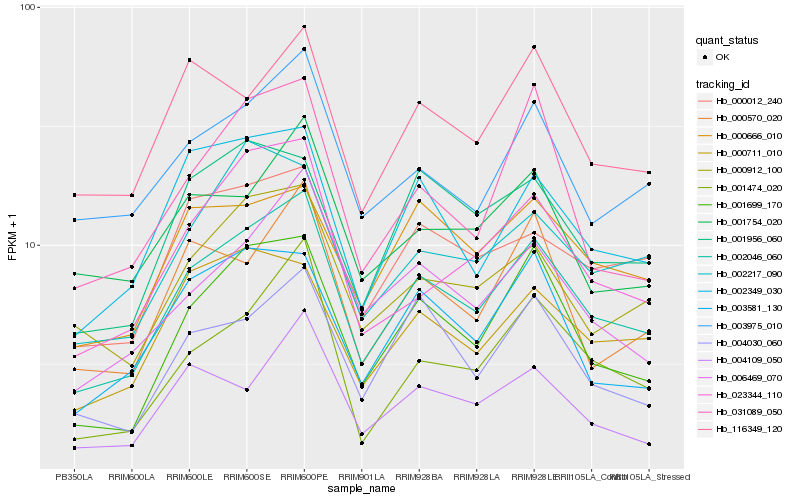
Hb_002046_060 |
0.0 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 2 |
Hb_001699_170 |
0.0852856359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002349_030 |
0.0952485961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000912_100 |
0.0960390254 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 5 |
Hb_006469_070 |
0.0962762329 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 6 |
Hb_003975_010 |
0.0985807268 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 7 |
Hb_000012_240 |
0.0985974464 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001474_020 |
0.1017110118 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 9 |
Hb_023344_110 |
0.1020668743 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 10 |
Hb_004030_060 |
0.1056305069 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 11 |
Hb_000711_010 |
0.1087313621 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 12 |
Hb_116349_120 |
0.1093826679 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_000570_020 |
0.1097118812 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 14 |
Hb_003581_130 |
0.1098290639 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_004109_050 |
0.1103021855 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 16 |
Hb_000666_010 |
0.1105308337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002217_090 |
0.1108292834 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 18 |
Hb_001956_060 |
0.1126672835 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 19 |
Hb_001754_020 |
0.11364507 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 20 |
Hb_031089_050 |
0.1165192771 |
- |
- |
kinase, putative [Ricinus communis] |