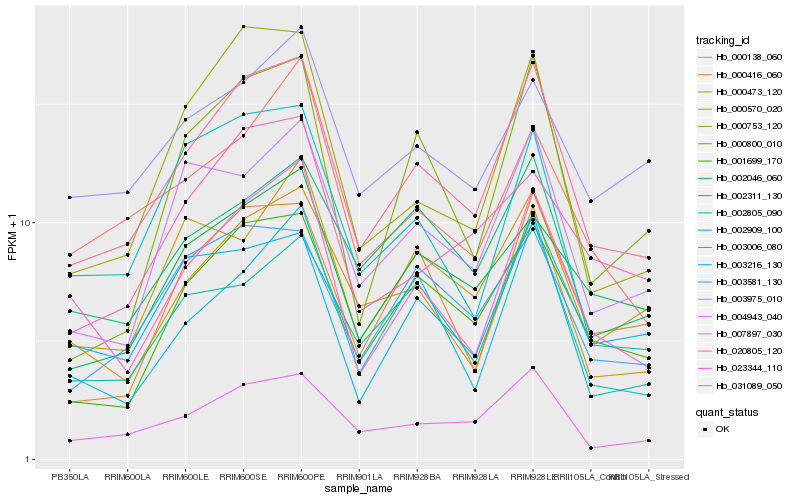
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031089_050 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_007897_030 |
0.0833045228 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |
| 3 |
Hb_001699_170 |
0.0904007657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000473_120 |
0.0981057753 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 5 |
Hb_020805_120 |
0.1051881099 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 6 |
Hb_003581_130 |
0.1092528529 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_004943_040 |
0.1125040204 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002311_130 |
0.1151579405 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002046_060 |
0.1165192771 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 10 |
Hb_000416_060 |
0.1171859013 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 11 |
Hb_000753_120 |
0.1174088408 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 12 |
Hb_002805_090 |
0.1200608506 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 13 |
Hb_003006_080 |
0.1230572698 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003216_130 |
0.1238998333 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 15 |
Hb_023344_110 |
0.1273528967 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 16 |
Hb_000800_010 |
0.1275029443 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 17 |
Hb_002909_100 |
0.1284667704 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 18 |
Hb_000138_060 |
0.128558424 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 19 |
Hb_003975_010 |
0.1290628382 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 20 |
Hb_000570_020 |
0.1294925546 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |