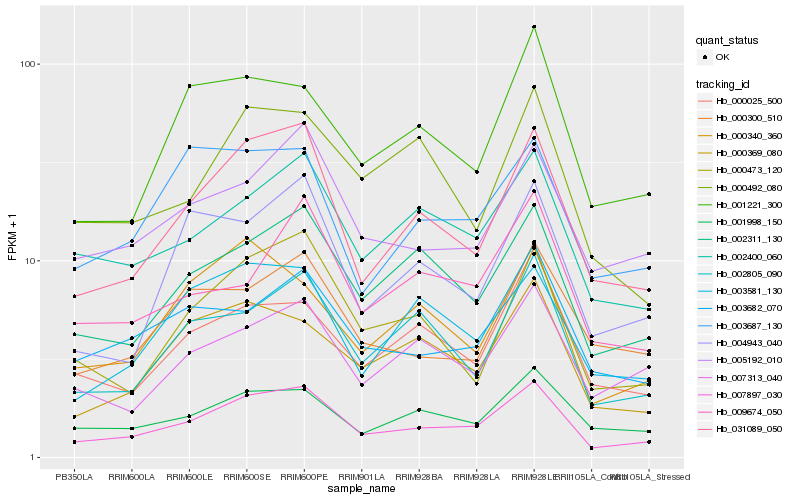
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007897_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |
| 2 |
Hb_031089_050 |
0.0833045228 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_002311_130 |
0.1040970922 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002400_060 |
0.1083731808 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 5 |
Hb_005192_010 |
0.1132900479 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 6 |
Hb_001998_150 |
0.1194918399 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 7 |
Hb_002805_090 |
0.12090159 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_000473_120 |
0.1219296439 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 9 |
Hb_003682_070 |
0.1231557844 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007313_040 |
0.1242221045 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 11 |
Hb_000369_080 |
0.1269502742 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 12 |
Hb_000025_500 |
0.1282345013 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 13 |
Hb_003581_130 |
0.1287579197 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001221_300 |
0.1319198136 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000300_510 |
0.1322158309 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 16 |
Hb_004943_040 |
0.1329627947 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003687_130 |
0.1351821584 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 18 |
Hb_000340_360 |
0.1358547356 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 19 |
Hb_000492_080 |
0.1370742865 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 [Jatropha curcas] |
| 20 |
Hb_009674_050 |
0.1371557697 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |