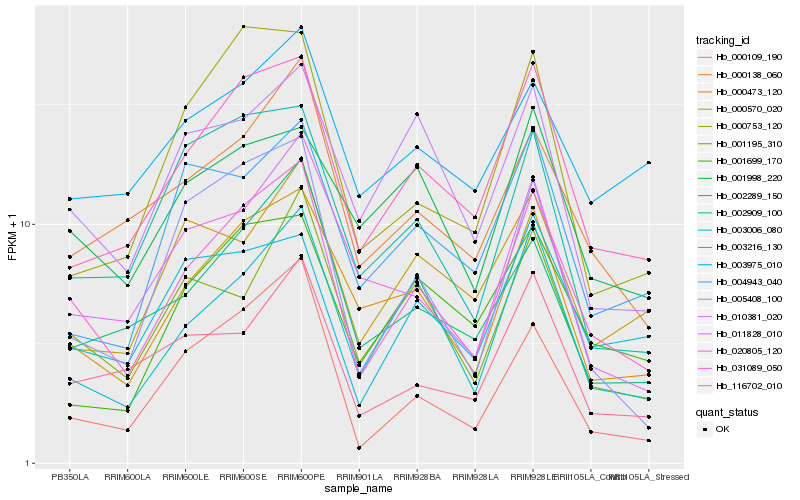
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020805_120 |
0.0 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 2 |
Hb_031089_050 |
0.1051881099 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_000473_120 |
0.1122347436 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 4 |
Hb_003006_080 |
0.122476448 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000138_060 |
0.1248601402 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 6 |
Hb_000109_190 |
0.1262840282 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 7 |
Hb_002289_150 |
0.1321759968 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 8 |
Hb_001195_310 |
0.134157853 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_116702_010 |
0.1354958864 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 10 |
Hb_005408_100 |
0.1385199848 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 11 |
Hb_011828_010 |
0.1402445535 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 12 |
Hb_002909_100 |
0.1435701948 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 13 |
Hb_003216_130 |
0.1440331807 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 14 |
Hb_010381_020 |
0.14687224 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 15 |
Hb_001998_220 |
0.1509521187 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000753_120 |
0.1514618898 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 17 |
Hb_003975_010 |
0.1515742509 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 18 |
Hb_004943_040 |
0.1519367251 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001699_170 |
0.1529383183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000570_020 |
0.1555904867 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |